Statistical CGP | Inductive CGP |
|---|
Scoring function | AUC | (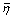 /η
max
) /η
max
) | Algorithm | AUC |
sens
| 0.634 | (1.2/1.8) |
NB
| 0.695 |
spec
| 0.464 | (0.8/1.5) |
LR
| 0.796 |
ppv
| 0.519 | (1.1/2.0) |
ADTree
| 0.780 |
npv
| 0.594 | (1.8/11.0) |
IBk
| 0.860 |
amss
| 0.578 | (2.4/96.6) |
J48
| 0.663 |
hmss
| 0.628 | (2.4/95.1) |
SMO/Poly
| 0.848 |
OR
| 0.537 | (1.2/2.3) |
SMO/RBF
| 0.782 |
chisq
| 0.767 | (3.2/109) | | |
bchisq
| 0.585 | (2.5/109) | | |
F
| 0.698 | (2.5/69.9) | | |
- Thirty-eight known genes were labelled as known (out of 4131 genes of the EC-K12 genome). The AUC in inductive CGP were calculated using stratified 10-fold cross-validation. Abbreviations: sens: sensitivity; spec: specificity; ppv: positive predictive value; npv: negative predictive value; amss: arithmetic mean of sensitivity and specificity; hmss: harmonic mean of sensitivity and specificity; OR: odds ratio; chisq: chi-square; bchisq: signed chi-square; F: F-measure; NB: naïve Bayes classifier; LR: logistic regression; ADTree: alternating decision tree; IBk: k-nearest neighbour classifier; J48: J48 decision tree; SMO: support vector machine trained by sequential minimal optimisation algorithm; Poly: polynomial kernel; RBF: radial basis function kernel.
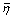 /η
max
)
/η
max
)