Figure 6
From: Structator: fast index-based search for RNA sequence-structure patterns

Construction of RNA secondary structure descriptors. (A) Non-overlapping alignment blocks of stem-loop regions excised from a multiple sequence-structure alignment and derived sequence-structure patterns. Since l
i
≤ r
i
<l
j
≤ r
j
and sequence regions S[l
i
... r
i
] fold into stem-loop structures for 1 ≤ i ≤ j ≤ 7, A = A1, A2, A3, A4, A5, A6, A7 is an ordered sequence of non-overlapping alignment blocks suitable to construct an RNA secondary structure descriptor  . The sequence-structure patterns
. The sequence-structure patterns 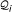 , i ∈ [1, 7] of
, i ∈ [1, 7] of  given on top of their underlying alignment blocks describe the seven marked stem-loops shown in the RNA secondary structure (B) of the Citrus tristeza virus replication signal (Rfam: RF00193). (C) Matches of RSSPs
given on top of their underlying alignment blocks describe the seven marked stem-loops shown in the RNA secondary structure (B) of the Citrus tristeza virus replication signal (Rfam: RF00193). (C) Matches of RSSPs 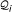 , i ∈ [1, 7], on sequence S, sorted in ascending order of their start position. (D) Graph-based representation of the matches of
, i ∈ [1, 7], on sequence S, sorted in ascending order of their start position. (D) Graph-based representation of the matches of 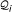 , i ∈ [1, 7]. An optimal chain of collinear non-overlapping matches is determined by computing an optimal path in the directed acyclic graph. Observe that not all edges in the graph are shown in this example and that the optimal chain (indicated here by their red marked members) is not necessarily the longest possible chain.
, i ∈ [1, 7]. An optimal chain of collinear non-overlapping matches is determined by computing an optimal path in the directed acyclic graph. Observe that not all edges in the graph are shown in this example and that the optimal chain (indicated here by their red marked members) is not necessarily the longest possible chain.