Figure 2

Performance of various similarity measures (noise-free case). (a) ROC curves obtained for the ID scoring scheme using the simple, conditional and partial Pearson correlation (μ
P
,  ,
,  ), where the diagonal of the cross-correlation matrix is set to 0. (b) ROC curves using the ID scoring scheme and different correlation coefficient, such as the simple Pearson correlation coefficient, where the diagonal of cross-correlation matrix is once 0 (μ
P
(diag 0)), and another time the diagonal is 1 (μ
P
(diag 1)). Furthermore, the ROC curves using the Spearman (μ
S
(diag 1)) and the Kendall (μ
K
(diag 1)) correlation coefficient, where the diagonal is 1 in both cases, are shown. (c) Evaluation of the ID scoring scheme using information-theoretic measures: simple, conditional and residual mutual information (μ
I
,
), where the diagonal of the cross-correlation matrix is set to 0. (b) ROC curves using the ID scoring scheme and different correlation coefficient, such as the simple Pearson correlation coefficient, where the diagonal of cross-correlation matrix is once 0 (μ
P
(diag 0)), and another time the diagonal is 1 (μ
P
(diag 1)). Furthermore, the ROC curves using the Spearman (μ
S
(diag 1)) and the Kendall (μ
K
(diag 1)) correlation coefficient, where the diagonal is 1 in both cases, are shown. (c) Evaluation of the ID scoring scheme using information-theoretic measures: simple, conditional and residual mutual information (μ
I
,  and
and  ). (d) Evaluation of the ID scoring scheme using measures based on symbolic dynamics: symbol sequence similarity (
). (d) Evaluation of the ID scoring scheme using measures based on symbolic dynamics: symbol sequence similarity (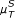 ), the mutual information of the symbol sequences (
), the mutual information of the symbol sequences (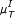 ) and the mean of these both (
) and the mean of these both (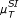 ), as well as the symbol sequence similarity of pairs of time points (
), as well as the symbol sequence similarity of pairs of time points (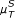 (pairs)) and the conditional entropy of the symbols obtained from the pairs of time points (
(pairs)) and the conditional entropy of the symbols obtained from the pairs of time points ( (pairs)). (e) The corresponding ROC curves illustrating the performance of the Time Shift scoring scheme using the Pearson correlation μ
P
, applied in addition to the CLR (measure: μ
S
) and the AWE (measure:
(pairs)). (e) The corresponding ROC curves illustrating the performance of the Time Shift scoring scheme using the Pearson correlation μ
P
, applied in addition to the CLR (measure: μ
S
) and the AWE (measure: 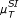 ) scoring scheme. (f) Performance of the AWE algorithm using the selected symbol based measures included in the this study, for example ROC curves for the symbol sequence similarity (
) scoring scheme. (f) Performance of the AWE algorithm using the selected symbol based measures included in the this study, for example ROC curves for the symbol sequence similarity (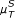 ), the mutual information of the symbol sequences (
), the mutual information of the symbol sequences (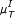 ), and the mean of these both (
), and the mean of these both (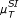 ).
).