Table 1 Feature ranking using (a) the correlation coefficient between input features and efficacy (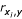 ), (b) mutual information feature selection (MIFS) with β = 0.75, (c) SVM-based Recursive Feature Elimination (SVM-RFE), and (d) best selection in [21] using the correlation coefficient.
), (b) mutual information feature selection (MIFS) with β = 0.75, (c) SVM-based Recursive Feature Elimination (SVM-RFE), and (d) best selection in [21] using the correlation coefficient.
From: Profiled support vector machines for antisense oligonucleotide efficacy prediction
FEATURE |
| FEATURE | MI (β = 0.75) | FEATURE | SVM-RFE ||W||2 | FEATURE |
| |
|---|---|---|---|---|---|---|---|---|
1 | ΔG | -0.35 | ΔG | 0.094 | ΔH | 0.680 | GGGA | 0.26 |
2 | # Cytosine | 0.31 | # Cytosine | 0.089 | ΔS | 0.671 | # Cytosine | 0.23 |
3 | TCCC | 0.28 | %GC content | 0.077 | ΔG | 0.193 | ΔH | -0.19 |
4 | 5pΔG | -0.26 | ΔG/length | 0.075 | # Cytosine | 0.045 | ΔG | -0.18 |
5 | ΔH | -0.24 | ΔH | 0.064 | Hairpin quality | 0.035 | CAGT | -0.18 |
6 | ΔH/length | -0.22 | ΔH/length | 0.061 | # Adenine | 0.024 | AGAG | 0.18 |
7 | %GC content | 0.22 | ΔS | 0.060 | # Thymine | 0.018 | GTGG | 0.17 |
8 | CCCT | 0.21 | # Adenine | 0.043 | Hairpin length | 0.014 | # Guanine | -0.15 |
9 | CCAC | 0.21 | # Guanine | 0.042 | 5pΔG | 0.009 | 3pΔG | 0.14 |
10 | CCCC | 0.21 | 5pΔG | 0.040 | 3pΔG | 0.005 | ΔS | -0.14 |
11 | CTCT | 0.20 | Hairpin quality | 0.027 | Dimer | 0.004 | CCCC | -0.13 |
12 | CCCA | 0.20 | Hairpin length | 0.024 | Hairpin energy (Mfold) | 0.003 | Hairpin quality | -0.11 |
13 | ACAC | -0.16 | Hairpin Energy | 0.022 | # Guanine | 0.001 | %GC content | 0.11 |
14 | # Adenine | -0.16 | # Thymine | 0.016 | Hairpin energy (vienna) | 0.000 | TGGC | -0.10 |
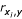 ), (b) mutual information feature selection (MIFS) with β = 0.75, (c) SVM-based Recursive Feature Elimination (SVM-RFE), and (d) best selection in [21] using the correlation coefficient.
), (b) mutual information feature selection (MIFS) with β = 0.75, (c) SVM-based Recursive Feature Elimination (SVM-RFE), and (d) best selection in [21] using the correlation coefficient.