Table 3 Extant scale normalization techniques used in this study. For a given microarray, if M
l
is a location-normalized log ratio, then M
s
is the scale-normalized log ratio, where M
s
= M
l
/ s, and s is median absolute deviation from the median (MAD), a robust estimate of the scale of the data distribution. The remaining abbreviations are as follows, median(M
l
): median value of M
l
values of spots on all microarrays in a data set; 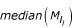 : median value of M
l
values of spots in PT group i on a microarray.
: median value of M
l
values of spots in PT group i on a microarray.
From: Evaluation of normalization methods for cDNA microarray data by k-NN classification
Name * | Description | Bioconductor R package/function (parameters) |
|---|---|---|
W SCALE | Within-microarray scale normalization M s = M l / s i | marrayNorm/maNormScale (norm="printTipMAD", subset = T, geo = T, Mscale = T) |
B SCALE | Between-microarray scale normalization s = median(M l - median(M l )) M s = M l / s | marrayNorm/maNormScale (norm="globalMAD", subset = T), geo = T, Mscale = T) |
WB SCALE | Step 1: Within-microarray scale normalization
| marrayNorm/maNormScale (norm="printTipMAD", subset = T, geo = T, Mscale = T) |
Step 2: Between-microarray scale normalization s = median(M l - median(M l ))
| marrayNorm/maNormScale (norm="globalMAD", subset = T, geo = T, Mscale = T) |
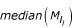 : median value of M
l
values of spots in PT group i on a microarray.
: median value of M
l
values of spots in PT group i on a microarray.

