Figure 3
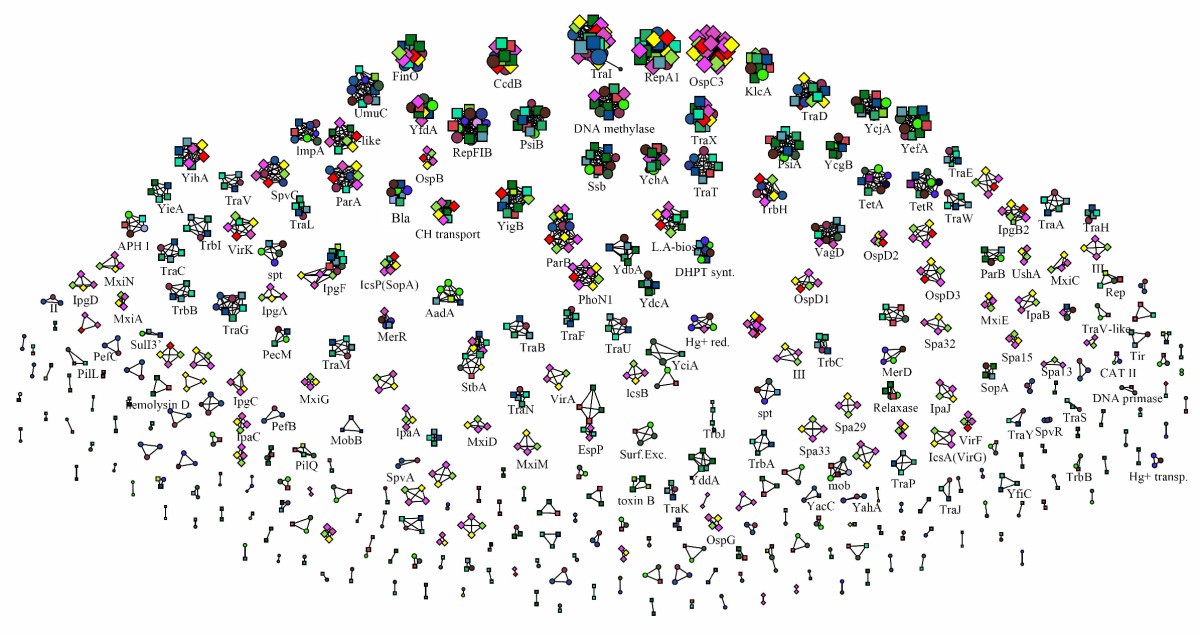
Uniform visualization of protein clustering. Uniform visualization of the similarity network for all of the 3701 proteins, displayed using a threshold identity for links of 40% (a degree of amino acid sequence identity sufficiently high to cluster together proteins that should perform the same function, and also allowing a better defined separation of all the main protein clusters [29, 30]). Groups of homologous proteins are separated, allowing the identification of proteins that very likely share an identical/similar function. The labels for some groups of proteins discussed in the text or very common are shown: KlcA, antirestriction protein involved in the broad-host range of IncP plasmids; FinO, RNA chaperone related to repression of sex pilus formation; CcdB, protein involved in plasmid stability by killing bacteria that lose the plasmid during cell division; TetA and TetR, proteins responsible for resistance to tetracycline; Bla, β-lactamases; AadA, and DHPT synthase, proteins involved in resistance to aminoglycosides or sulfonamides, respectively. Tra and Trb, proteins requested for sex pilus formation; Mxi, Spa, Ipa, Ipg and Osp, proteins that are part of the type III secretion system.