Fig. 7
From: Magic-BLAST, an accurate RNA-seq aligner for long and short reads
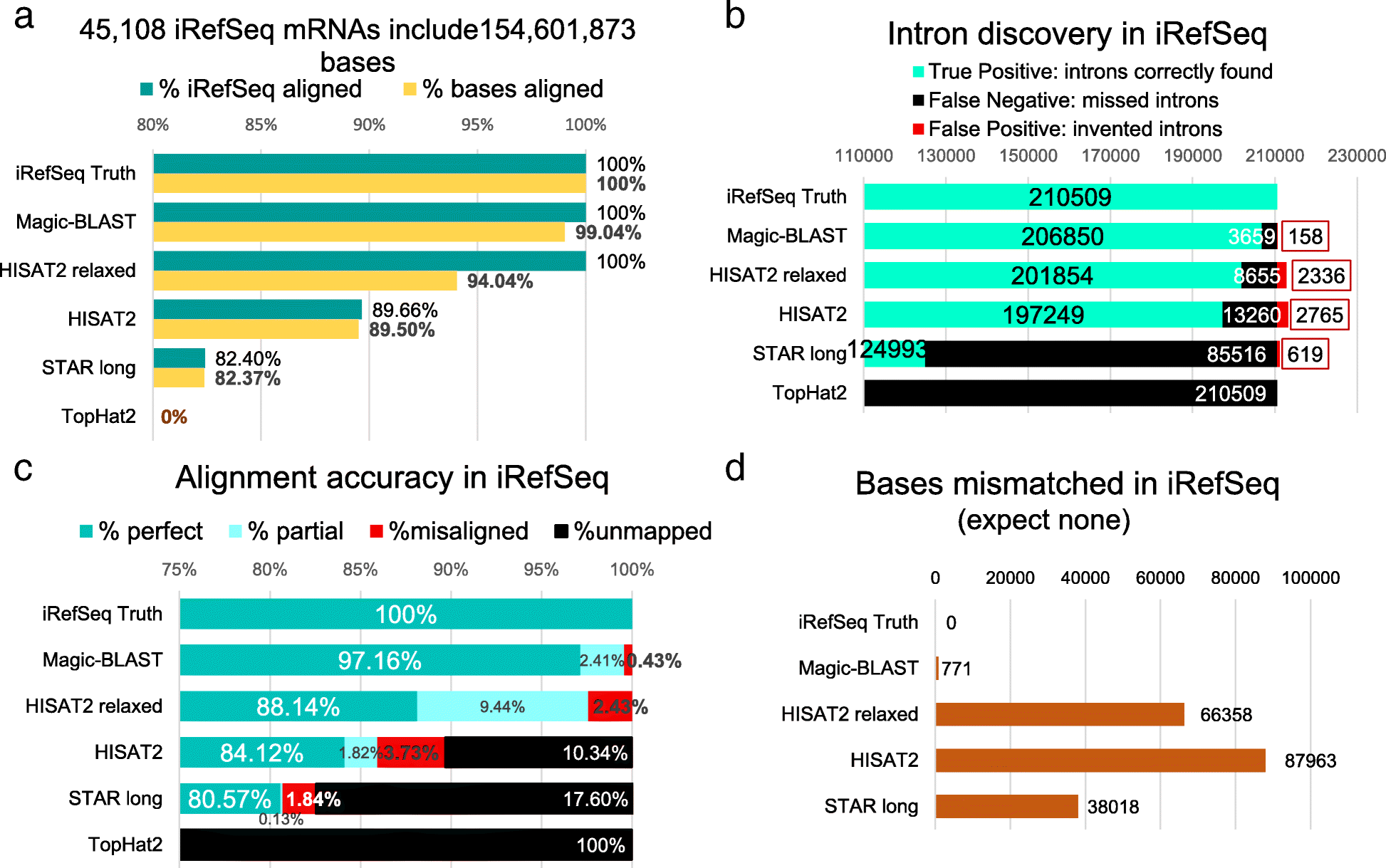
Characteristics of alignments of the iRefSeq set. a The 45,108 iRefSeq include 154,601,873 bases exactly matching the genome (Truth). For each program, the percentage of iRefSeq sequences aligned (green) and the percentage of bases aligned (yellow) are given. In case of multiple alignments, each read contributes only once, at its primary position. Note that the scale is from 80 to 100%. b Intron discovery in iRefSeq: the number of introns correctly found (green, TP), the number present in the truth but missed by the aligner (black, FN) and the number of invented introns, found by the aligner but absent from the truth (red, FP) are shown. c Accuracy of the alignments: the true alignment of each read is defined by the RefSeq annotation (GRCh38 GFF file). A read is considered exactly aligned (green) if it is completely aligned, without mismatch, and starts and ends at the same chromosomal coordinates as the truth. A partial (light blue) must be within the true alignment. For misaligned reads (red), at least part of the alignment does not overlap the truth. Unmapped reads are shown in black. Note that the scale is from 75 to 100%. d The number of mismatches reflects mis-mappings, since by construction the iRefSeq exactly match the genome