Fig. 1
From: ARPIR: automatic RNA-Seq pipelines with interactive report
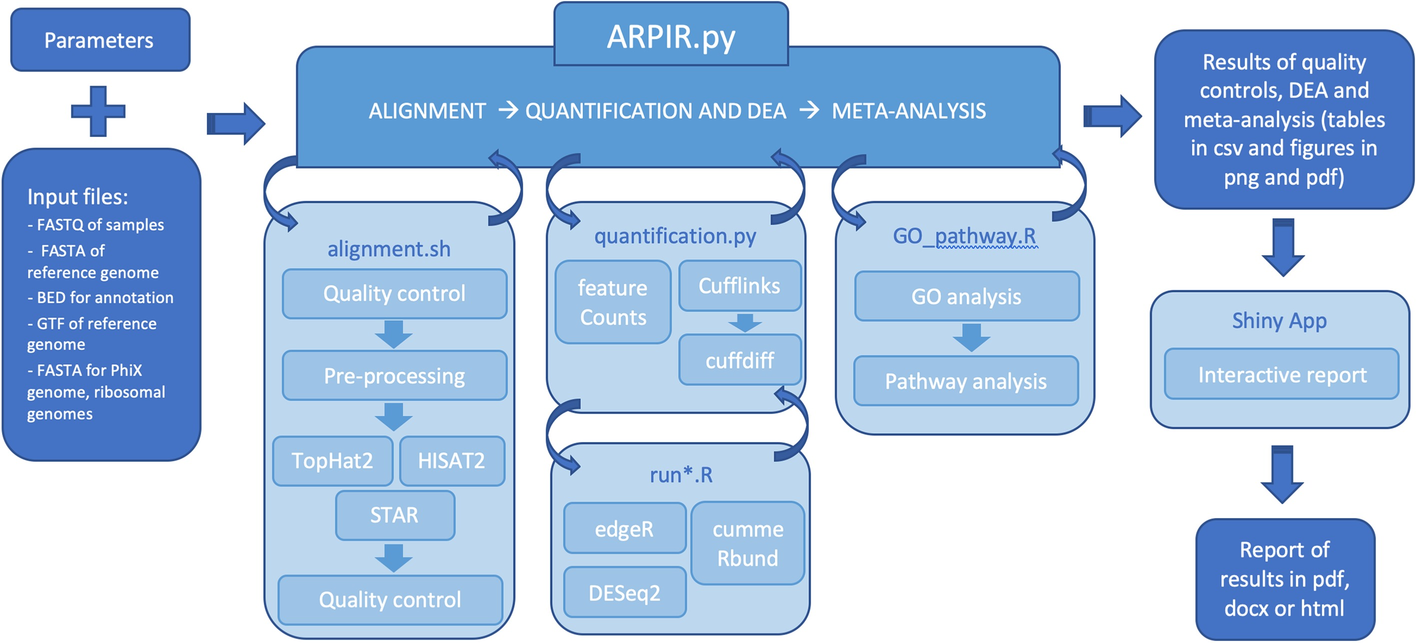
Workflow of ARPIR pipeline. The ARPIR pipeline, starting from the input files and parameters, performs an RNA-Seq analysis. First of all the primary-analysis, a quality control on the FastQ files occurs, followed by a pre-processing and alignment, which can be done through TopHat2, HISAT2 or STAR, finally there is a new quality control on the BAM files. The secondary-analysis is the quantification and differential analysis, which can follow the featureCounts-edgeR, featureCounts-DESeq2 or Cufflinks-cummeRbund pipelines. Then an optional tertiary-analysis follows, composed of a GO analysis and a Pathway analysis. The results obtained can then be viewed in a Shiny App and possibly downloaded to a report