Fig. 1
From: scSemiAAE: a semi-supervised clustering model for single-cell RNA-seq data
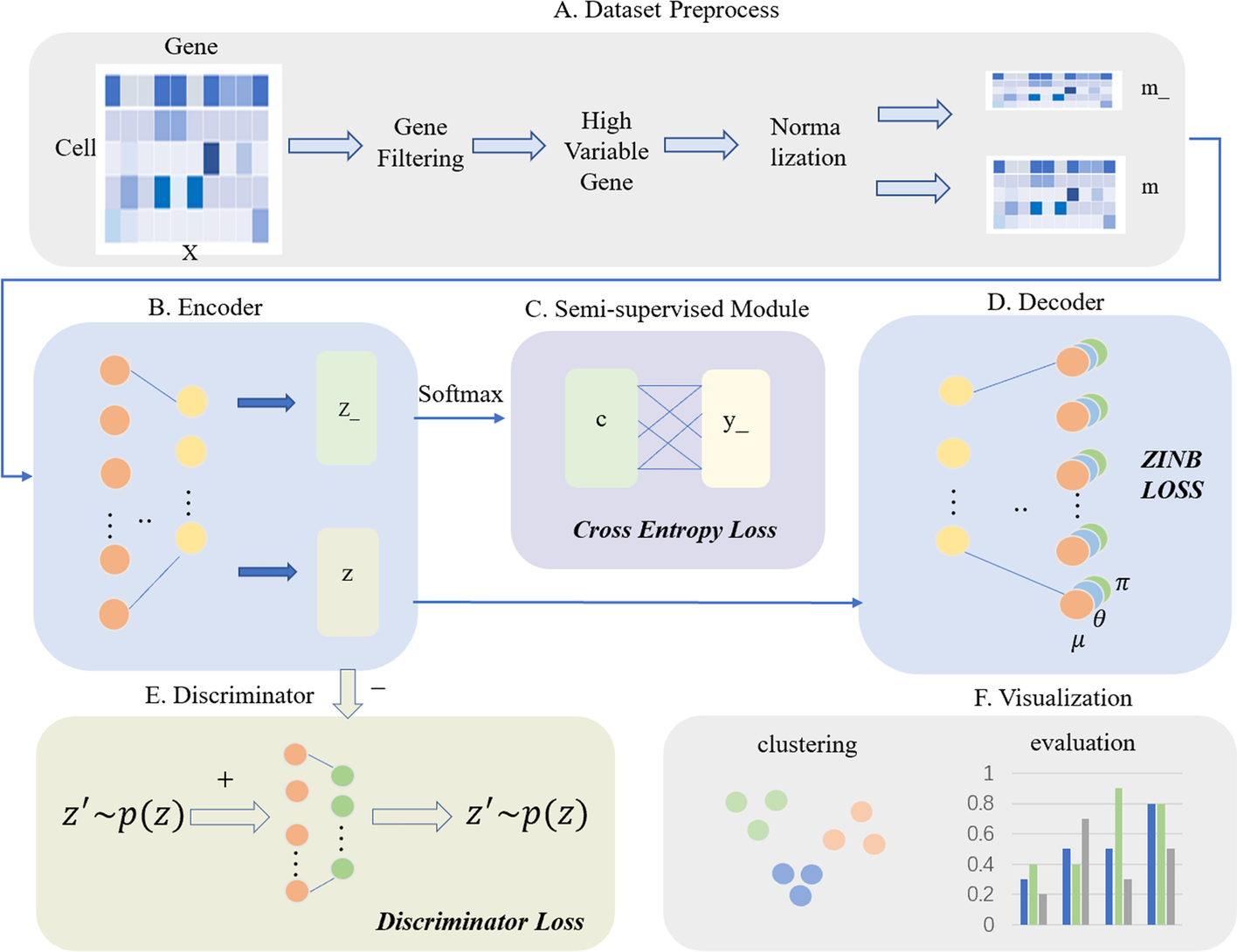
The illustration of scSemiAAE model. A The scRNA-seq count matrix X is preprocessed through gene filtering, screening of highly variable genes, and normalization. Next, it is divided into m_ and m depending on whether it contains true labels. B The encoder receives m_ and m to generate the corresponding latent variables z_ and z, respectively. C The SoftMax layer transforms the latent vector z_ into the pseudo-label c, which is then combined with the partial true label y_ to create a cross-entropy loss. D The decoder reconstructs the potential representation z with a zero-inflated negative binomial loss constraint. E Simultaneously, the latent feature z is fed to the discriminator for adversarial training, comprising the discriminator loss. F After completing training process, all the latent z and labels c are concatenated, and the final clustering results are given by a Gaussian mixture model