Table 1 Measures
MEASURE | "SIMPLE" (PAIRWISE) | CONDITIONAL | PARTIAL |
|---|---|---|---|
Euclidean distance | μ EC , [63] | - | - |
Ls Norm (here s = 10) | μ L , [64] (in literature s = 3) | - | - |
Manhattan distance | μ MA , [64] | - | - |
dynamic time warping distance | μ W , [43] | - | - |
Pearson's correlation | μ P , [65] |
|
|
Spearman's correlation | μ S , [66] | - | - |
Kendall's correlation | μ K , [67] | - | - |
mutual information | μ I , [66] |
|
|
coarse-grained information rate |
|
| - |
Granger causality index | μ G , [68] |
|
|
symbol sequence similarity |
| - | - |
mutual information of symbol sequence |
| - | - |
mean of symbol sequence similarity and μI |
| - | - |
conditional entropy of symbol sequence | - |
| - |
 , [
, [ , [
, [ , [
, [ (new)
(new) , [
, [ , [
, [ , [
, [ , [
, [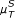 , [
, [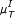 , [
, [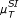 , [
, [ , [
, [