Figure 4
From: MUSCLE: a multiple sequence alignment method with reduced time and space complexity
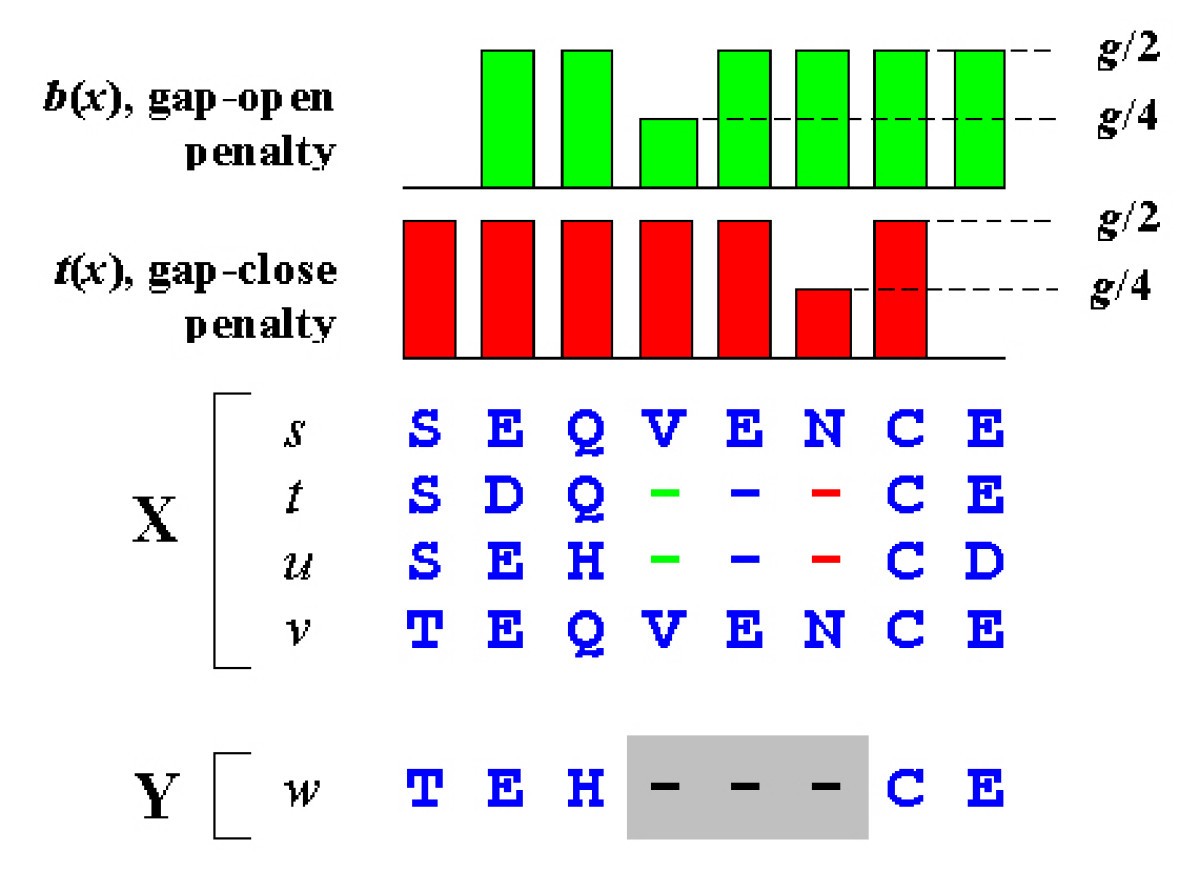
Position-specific gap penalties. An alignment of two profiles X and Y. Gaps in sequences t and u are embedded in X. Y contains a single sequence w. The gap in w (gray background) is inserted to align the profiles and is not part of Y. Consider the SP score for this alignment. We need not consider pairs of sequences in X as their scores are unchanged under all possible alignments of X to Y, leaving the inter-profile pairs (s, w), (t, w), (u, w) and (v, w). Note that there is no gap penalty for the pairs (u, w) and (v, w) as these pairs do not have gaps relative to each other. The remaining pairs (t, w) and (u, w) are assessed a penalty g + 3e for the gap in Y. The total over all pairs of open or close penalties due to a gap in Y is thus reduced in proportion to the fraction of sequences in X having a gap with the same open or close position. We incorporate this into the PSP score by using position-specific gap penalties b(x) and t(x). For example, b(x) in column 4 of X is half the default value because half of the sequences in X open a gap in that column. Note that there is no open penalty at the N-terminal and no close penalty at the C-terminal. This causes terminal gaps to receive half the penalty of internal gaps.