Table 1 Notations.
From: An integrated Bayesian analysis of LOH and copy number data
Het | heterozygous |
|---|---|
Hom | homozygous |
NHet | not heterozygous (is used when we cannot distinguish between two equal nucleotides, i.e. homozygosity, and the loss of one copy) |
| {Het, Hom} |
| {ø, Het, Hom} |
| {Het, NHet, NoCall} |
X N | true genotypes in normal cells ( |
X | true genotypes in abnormal cells (X
i
∈ |
Y | genotypes detected by the genotype calling algorithm (Y
i
∈ |
Y cn | raw copy number data |
| copy number events/aberrations |
| occurrence of copy-neutral LOH (i.e. IBD/UPD event) |
| IBD/UPD & copy number aberrations ({ |
cn | all copy number information (both raw data and estimated profile by mBPCR) |
p | vector of posterior probabilities to be a breakpoint (for all SNP positions) |



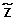


 = w} = {
= w} = { ,
,  = u} for some w, z, u)
= u} for some w, z, u)