Fig. 1
From: Internal and external normalization of nascent RNA sequencing run-on experiments
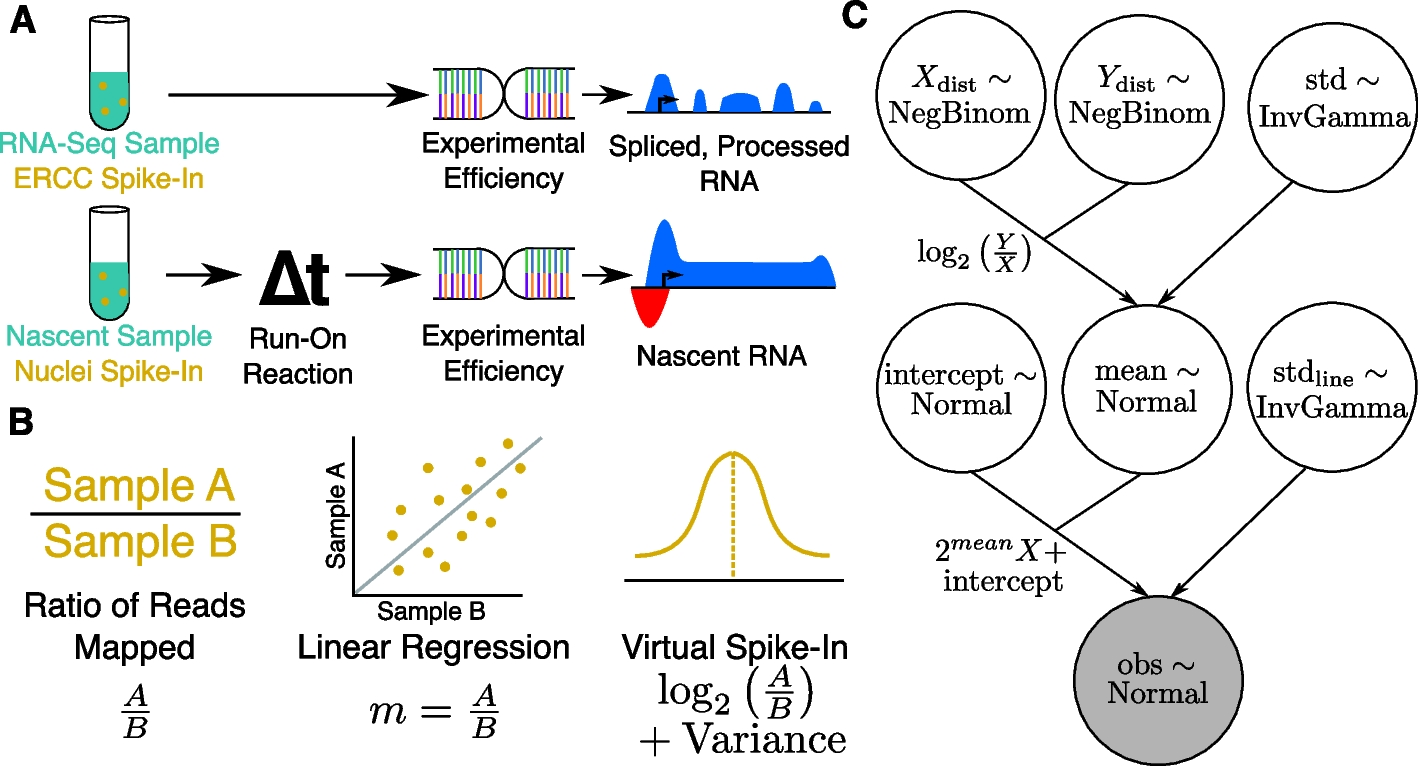
A Bayesian model describing normalization data for nascent RNA sequencing data. A Schematic showing typical external control, handling, and resulting data profile differences between RNA-seq (top) and run-on nascent RNA sequencing assays (bottom). Note that run-on efficiency is assumed to be equivalent between spike-in nuclei and experimental nuclei. B Quantifying a normalization factor is accomplished either by a naive ratio of total reads approach (left), linear regression (middle), or by the Bayesian model proposed here (right). Linear regression (middle) is more resistant to noise and outliers, but does not provide a reliable way to measure the variance of the normalization estimate. The Bayesian model (right) converts the slope \(m = \frac{A}{B}\) to \(\log\) space, converting the multiplicative nature of the normalization factor to a linear one, for which normalization factors can be readily inferred as a normal distribution with variance. C A plate diagram showing the VSI model as implemented in pymc3. Briefly, we estimate our count distributions X and Y (top row) with a negative binomial. The ratio of two negative binomial distributions is approximately log-normal, so we derive a normal distribution called mean (middle) as the log of the ratio of Y and X with some variance (top right), estimated as an inverse gamma distributed random variable. With the estimation of the mean established, we then add additional parameters to describe the intercept, and variance of the actual line of best fit. This is done so that the parameter mean is estimating an error in log-transformed space, as discussed in Panel (B)