Fig. 1
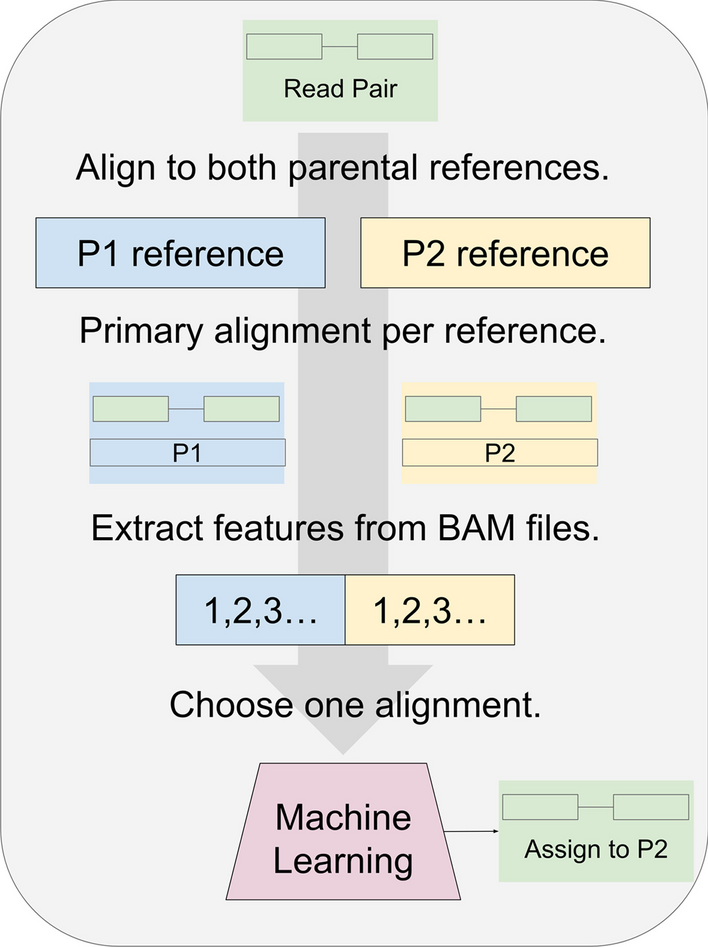
Process for classifying RNA-seq by machine learning. For training the model, read pairs are obtained from parents P1 and P2. Each read pair is aligned to the P1 and P2 references separately. A filter removes all but the primary alignment per read pair. The process can use transcriptomes or genomes as references, and any suitable aligner. For read pairs that align to both P1 and P2, features are extracted and given to the machine learning model. Initially, the model is given the true parent label per read pair and trained to predict this. After training, read pairs from the hybrid cross are given instead. In this illustration, after a hybrid read pair (green) is aligned to the P1 (blue) and P2 (yellow) references, the trained model (red) chooses P2 as the more likely source. For most of this study, the model was a random forest binary classifier. Read pairs classified by this process could be binned by gene and quantified to detect allele-specific gene expression in the hybrid