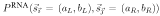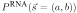- Methodology article
- Published:
Noncoding RNA gene detection using comparative sequence analysis
BMC Bioinformatics volume 2, Article number: 8 (2001)
Abstract
Background
Noncoding RNA genes produce transcripts that exert their function without ever producing proteins. Noncoding RNA gene sequences do not have strong statistical signals, unlike protein coding genes. A reliable general purpose computational genefinder for noncoding RNA genes has been elusive.
Results
We describe a comparative sequence analysis algorithm for detecting novel structural RNA genes. The key idea is to test the pattern of substitutions observed in a pairwise alignment of two homologous sequences. A conserved coding region tends to show a pattern of synonymous substitutions, whereas a conserved structural RNA tends to show a pattern of compensatory mutations consistent with some base-paired secondary structure. We formalize this intuition using three probabilistic "pair-grammars": a pair stochastic context free grammar modeling alignments constrained by structural RNA evolution, a pair hidden Markov model modeling alignments constrained by coding sequence evolution, and a pair hidden Markov model modeling a null hypothesis of position-independent evolution. Given an input pairwise sequence alignment (e.g. from a BLASTN comparison of two related genomes) we classify the alignment into the coding, RNA, or null class according to the posterior probability of each class.
Conclusions
We have implemented this approach as a program, QRNA, which we consider to be a prototype structural noncoding RNA genefinder. Tests suggest that this approach detects noncoding RNA genes with a fair degree of reliability.
Introduction
Some genes produce functional noncoding RNAs (ncRNAs) instead of coding for proteins [1, 2]. For protein-coding genes, we have computational genefinding tools [3] that predict novel genes in genome sequence data with reasonable efficiency [4]. For ncRNA genes, there are as yet no general genefinding algorithms. The number and diversity of ncRNA genes remains poorly understood, despite the availability of many complete genome sequences. Gene discovery methods (whether experimental or computational) typically assume that the target is a protein coding gene that produces a messenger RNA.
New noncoding RNA genes continue to be discovered by less systematic means, which makes it seem likely that a systematic RNA genefinding algorithm would be of use. Recent discoveries have included RNAs involved in dosage compensation and imprinting [5], numerous small nucleolar RNAs involved in RNA modification and processing [6–8], and small riboregulatory RNAs controlling translation and/or stability of target mRNAs [9, 10]. Mutations in the gene for RNase MRP are associated with cartilage-hair hypoplasia (CHH), a recessive pleiotropic human genetic disorder [11]. The CHH locus eluded positional cloning for some time; the RNase MRP gene was only detected in the completely sequenced CHH critical region because the RNase MRP sequence was already in the databases.
We have previously explored one RNA genefinding approach with very limited success [12]. Maizel and coworkers [13–15] had hypothesized that biologically functional RNA structures may have more stable predicted secondary structures than would be expected for a random sequence of the same base composition. Though we could confirm some anecdotal results where this was true, we were forced to the conclusion that in general, the predicted stability of structural RNAs is not sufficiently distinguishable from the predicted stability of random sequences to use as the basis for a reliable ncRNA genefinding algorithm. Nonetheless, conserved RNA secondary structure remained our best hope for an exploitable statistical signal in ncRNA genes. We decided to consider ways of incorporating additional statistical signal using comparative sequence analysis.
We were motivated by the work of Badger & Olsen [16] for bacterial coding-region identification. Badger & Olsen use the BLASTN program [17] to locate genomic regions with significant sequence similarity between two related bacterial species. Their program, CRITICA, then analyzes the pattern of mutation in these ungapped, aligned conserved regions for evidence of coding structure. For example, mutations to synonymous codons get positive scores, while aligned triplets that translate to dissimilar amino acids get negative scores. (CRITICA then subsequently extends any coding-assigned ungapped seed alignments into complete open reading frames.)
Here we extend the central idea of the Badger & Olsen approach to identify structural RNA regions. Our extensions include: (1) using fully probabilistic models; (2) adding a third model of pairwise alignments constrained by structural RNA evolution; (3) allowing gapped alignments; and (4) allowing for the possibility that only part of the pairwise alignment may represent a coding region or structural RNA, because a primary sequence alignment may extend into flanking noncoding or nonstructural conserved sequence. These extensions add complexity to the approach. We use probabilistic modeling methods and formal languages to guide our construction. We use "pair hidden Markov models" (pair-HMMs) (introduced in [18]) and a "pair stochastic context free grammar" (pair-SCFG) (a natural extension of the pair-HMM idea to RNA structure) to produce three evolutionary models for "coding", "structural RNA", or "something else" (a null hypothesis). Given three probabilistic models and a pairwise sequence alignment to be tested, we can calculate the Bayesian posterior probability that an alignment should be classified as "coding", "structural RNA", or "something else".
Our approach is designed to detect conserved structural RNAs. Some ncRNA genes do not have well-conserved intramolecular secondary structures, and some conserved RNA secondary structures function as cis-regulatory regions in mRNAs rather than as independent RNA genes. We will be using the term "ncRNA gene" to refer to our prediction targets, but it must be understood that this really means a conserved RNA secondary structure that may or may not turn out to be an independent functional ncRNA gene upon further analysis.
Algorithm
Overview of the approach: simple, ungapped global case
The key idea is to produce three probabilistic models (RNA, COD, and OTH) describing different evolutionary constraints on the pattern of mutations observed in a pairwise sequence alignment. We will first introduce toy versions of these models, for clarity.
All three models use the "pair-grammar" formalism described in [18]. A standard hidden Markov model (HMM) generates a single observable sequence by emitting single residues, whereas a pair-HMM generates two aligned sequences X, Y by emitting a pair of aligned residues at a time (or single residues in either sequence to deal with insertion and deletion).
The OTH model assumes mutations occur in a simple position-independent fashion. OTH has 4 × 4 core parameters, which are the pairwise alignment probabilities POTH(a, b) – that is, the joint probabilities of emitting an alignment of a nucleotide a in sequence X and a nucleotide b in sequence Y (Table 1). OTH represents our null hypothesis. The probability of the alignment given the OTH model is just the product of the probabilities of the individual aligned positions.
The COD model assumes that the aligned sequences encode homologous proteins. In a coding region, we intuitively expect to see mutations that make conservative amino acid substitutions; in particular, we expect an abundance of synonymous mutations. To capture this, COD has 64 × 64 core parameters, which are PCOD (a1a2a3, b1b2b3), the probabilities of the correlated emission of two codons – that is, three nucleotides a1a2a3 in sequence X, aligned to three nucleotides b1b2b3 in sequence Y. (See Table 1 for an example of pair codon probabilities.) The probability of the alignment given the COD model for a particular reading frame is the product of the probabilities of the individual aligned codons in that frame. Since we don't know the correct frame a priori, the overall probability of an alignment  is a sum over all six frames f,
is a sum over all six frames f,

and we assume that all frames are a priori equiprobable in the alignment (P(f|COD) =  ).
).
The RNA model assumes that the pattern of mutation significantly conserves a homologous RNA secondary structure. Intuitively, we expect a significant abundance of pairwise-correlated mutations that preserve Watson-Crick complementarity in an (as yet unknown) structure. To capture this, the core parameters in RNA are the 16 × 16 probabilities PRNA(a
L
a
R
, b
L
b
R
) – that is, the probabilities associated with the correlated emission of one base-pair (a
L
a
R
) in sequence X aligned to a homologous base-pair (b
L
b
R
) in sequence Y (Table 1). Single stranded positions in the alignment are modeled by PRNA(a, b), the same functional form as in the OTH model. For a given alignment  of known structure s, the probability P(
of known structure s, the probability P( |s, RNA) is a product of terms PRNA(x
i
x
j
, y
i
y
j
) for all base paired positions i, j and PRNA(x
k
, y
k
) for all single stranded positions k in the alignment. Since we don't know the correct structure a priori, the overall probability of an alignment
|s, RNA) is a product of terms PRNA(x
i
x
j
, y
i
y
j
) for all base paired positions i, j and PRNA(x
k
, y
k
) for all single stranded positions k in the alignment. Since we don't know the correct structure a priori, the overall probability of an alignment  given by the RNA model is a sum over all structures s:
given by the RNA model is a sum over all structures s:

But here, we cannot assume equiprobability for the various structures s as we did for coding frames f above; in fact, calculating P(s|RNA) implies a full probabilistic model describing favorable and unfavorable RNA secondary structures. The necessary machinery for calculating this weighted sum is exactly what we developed previously for searching for significant structure in single sequences [12]. In that paper we parameterized a stochastic context-free grammar (SCFG) that incorporates a model of hairpin loops, stems, bulges, and internal loops, including stacking and loop-length distributions, making a probabilistic counterpart for the widely used MFOLD program for RNA structure prediction. The SCFG we use here is almost the same, with the difference that now we generate two aligned sequences simultaneously: i.e., a pair-SCFG. The summation over all possible structures can be done efficiently using an SCFG Inside algorithm (a dynamic programming algorithm).
In Figure 1 we present an example of three different alignments with different mutation patterns, and how they would be scored with the three different models.
Three pairwise alignments of identical composition with identical number of base substitutions can be classified by distinctive patterns of mutation caused by different selective constrains: the position-independent null hypothesis (top), a coding region (middle), or a structural RNA (bottom). We indicate how each alignment is scored according to the model that best fits the pattern of mutations: one position at the time for OTH, one codon at the time for COD (integrated over all six possible frames), and as a combination of base-paired positions and single positions for RNA (integrated over all possible secondary structures).
Finally, in order to classify the input alignment as RNA, COD, or OTH, we use the three likelihoods to calculate a Bayesian posterior probability, under the simple assumption that the three models are a priori equiprobable. Alignments with high RNA posterior probabilities are interpreted as candidate ncRNA genes.
For scoring purposes, it will also be useful to calculate log-odds scores in the standard manner [19] relative to a fourth model, which we will call IID. In IID, we assume the two sequences are nonhomologous independent, identically distributed sequences. The IID model has 8 parameters corresponding to the expected base compositions ofthe two sequences, PX(a) and PY(b).
Parameter estimation in the simple case
Parameter estimation is crucial for our approach. The three models have to be calibrated to an overall similar evolutionary divergence time, and to similar base compositions. Else, one model might be artifactually favored over the others because of the degree of conservation or the base composition in an input alignment, not because of the pattern of mutation.
In an ideal world, we could empirically estimate the parameters of each model using training sets of pairwise alignments culled from real RNAs, coding regions, and other conserved noncoding regions, using pairwise alignments that were all about the same percent identity. Unfortunately it is unlikely that we can amass suitably large training sets. Instead, we take a somewhat ad hoc approach that ties the parameters of all three models as much as possible to a particular choice of a standard amino acid substitution matrix, such as BLOSUM62. We will derive joint codon probabilities from the chosen scoring matrix, then use these codon probabilities to calculate the average single nucleotide substitution probabilities in OTH, then combine these OTH substitution parameters with base-pair frequencies to obtain the parameters of the RNA model. This procedure is as follows.
First the 64 × 64 codon emission probabilities PCOD(a1a2a3,b1b2b3 |t), for some divergence "time" t, are derived from the chosen substitution matrix (i.e. the choice of matrix defines t). We make an independence assumption that the conditional probability of each codon depends only on its own encoded amino acid – i.e., it does not depend on the the other codon – so we can use the approximation

for a1, a2, a3, b1, b2, b3 ∈ {A, C, G, U} and A, B ∈ {amino acids}. (An example of where this independence assumption is violated: for equiprobable codon bias, our parameters will say that aligning TCA to AGT is as likely as aligning TCA to TCG because all three are Ser codons, despite the fact that the first case requires three transversions.) P(A, B|t) are the joint target probabilities of aligned amino acids obtained from the amino acid score matrix, such as BLOSUM62 [20], as described by [19]. The terms P(a1a2a3|A) are the probabilities of observing a particular codon given a particular amino acid; these terms can include a codon-bias model [21] and, if desired, a substitution error model to deal with error-prone sequence data. The sum over all possible amino acids in equation (3) is relevant only when a substitution error model applies, since otherwise each observed codon can only mean one possible amino acid.
The 16 mutation probabilities for the OTH model are then obtained by marginalizing the corresponding codon-codon emission probabilities in equation (3), in the following way:

The 16 × 16 core parameters of the RNA model are calculated by combining the OTH model (which sets the average divergence of the two sequences) with some additional parameters that specify the probability of base pairs. This involves making an independence assumption:

Alternatively, we can symmetrically derive a equation in which the divergence is controlled by the mutation probability of the right position instead of the left position. We calculate the overall joint probability of the aligned base pairs as the average of these two equations:


Here Ppair(a L a R |t), Ppair(b L b R |t) are just the probabilities of the various sorts of base pairs (GC, AU, GU) in a single RNA structure.
Extension of the models to gapped local alignments
In order to deal with gapped local alignments (as reported by BLASTN, for instance), we will have to extend the models to deal with two problems.
Obviously we have to deal with the presence of insertions and deletions (indels) in the alignments. In fact, there is information in the indels that we would like to capture. Indels in coding sequence will occur in multiples of three nucleotides to preserve coding frame. The length of an RNA stem may vary in two homologous structures, leading to long-distance correlated indels.
We also have to recognize that the bounds of reported local sequence alignments will not usually correspond to the true bounds of a functional coding or RNA sequence. It is therefore too simplistic to assume that all the residues in the alignment should be assigned to a single choice of model. For example, Figure 3 shows a real BLASTN alignment containing a U2 small nuclear RNA gene conserved between Caenorhabditis elegans and Caenorhabditis briggsae; the alignment extends beyond the U2 structural RNA into less conserved flanking noncoding sequences.
Alignment generated by WUBLASTN between C. elegans clone F08G2 and C. briggsae clone G42J05. This alignment contains a U2 snRNA gene. Underlined is the actual U2 gene (coordinates: 3128–3313). The secondary structure (placed above the alignment) is provided by [43] and includes a pseudoknot. We observe four compensatory mutations (represented with "*") which conserve the secondary structure of the U2 gene.
Both of these problems can be addressed using the "pair-grammar" formalism introduced by [18]. A pair-HMM for the OTH model that can generate insertions is shown in Figure 2. State XY emits two aligned nucleotides simultaneously in both sequences with probability POTH(a, b|t), while there is also a non-null probability of moving to states X or Y that generate nucleotides in only one of the two sequences, and gaps in the other one.
In order to make the alignments local, we add flanking states to the models. These flanking states allow us to score portions of the alignments as if they were unaligned residues that are unassigned to the model. The IID HMM, which emits both sequences independently, is composed solely of these flanking states (Figure 4).
The OTH Model
The complete OTH model, a pair hidden Markov model, is diagrammed in Figure 5. The flanking double-circled states F L , F R , and F J are a shorthand for a full IID model of the type in Figure 4, which allow the alignment to be flanked or interrupted by runs of unassigned (independent) residues. (In general we will use the convention that single-circled states are "single states", and double-circled states represent some "composite state" of some kind previously defined. This differs from a convention in formal languages in which double-circled states are terminal states of a finite-state automaton [22].)
Description of the probabilistic OTH model for local gapped alignments. This model permits the local alignment of two sequences. The flanking states F L , F R , F J with double circles represent composite states defined as in Figure 4.
The OTH model requires us to specify emission probabilities for the state XY (that emits two aligned nucleotides), and also for the X and Y states (that emit one nucleotide "aligned" to a gap character in the other sequence). The emission probabilities for state XY, PXY (a, b|t), are simply the mutation probabilities POTH(a, b|t) of the toy ungapped OTH model, as described above. The emission probabilities for states X and Y are obtained by marginalization of the PXY's:

The COD Model
The complete COD model, a pair hidden Markov model, is diagrammed in Figure 6. A new degree of "locality" is introduced. In addition to regions of the alignment that are better left "unaligned" (i.e. generated by the flanking states of an IID model), we want to model regions of the alignment that are not coding but still well-conserved. To model this, we add three full copies of OTH models to the core of the COD model, indicated by the symbols O B , O E , and O J . We represent a full OTH model with:
Description of the probabilistic COD model for local gapped alignments. The double-circled states O L , O R , O J (defined as in Figure 5) represent composite states responsible for possible independently aligned emissions within the COD model.

with the understanding that any arrow that goes into "O" indicates a transition into the "S FL " state of the F L flanking model, and any arrow leaving "O" emerges from the "T FR " state of the F R flanking model. In this way the COD model can score a coding-aligned region that is nested between other independently-aligned regions.
The different COD states described in Figure 6 emit correlated codon pairs, possibly with indels. To deal with BLASTN misalignments of codons and possible applications to error-prone sequence data (expressed sequence tags or low-pass genome shotgun), we model -1 or +1 frameshifts (by having a probability of emitting abnormal codons of 2 or 4 nucleotides), in addition to the more expected indels of multiples of three nucleotides. (No explicit transition for C E → C B is necessary; the intermediate sub-model "O J " has a non-emitting path that deals with consecutive codons.)
Codon emission probabilities for the different coding states are derived from the joint codon probabilities PCOD given in equation (3) for the toy case. For incomplete codons we do the convenient marginalizations. For example,

Notice that there are three different C(3, 2) states, of which we have only described one in equation (9). Similarly there are four different C(3, 4) states, and six different C(2, 4) states, depending on the position of the gaps. We will represent these codon-emission probabilities in general by Pα, β (a1...aα, b1...bβ) with α, β = {0, 2, 3, 4} and a, b ∈ {A, C, G, U}.
The RNA Model
The complete RNA model, a pair stochastic context free grammar (pair-SCFG), is crudely diagrammed in Figure 7. The crucial SCFG machinery of the model is encapsulated in the RNA state of the diagram. This pair-SCFG, similar to the SCFG described in [12], has three states labeled W, W B , V. They correspond to the V and W dynamic programming matrices in [23], and to the matrices wx, wbx and vx of the diagrammatic representation in [24, 25]. We use the diagrams as a convenient visual representation to enumerate the configurations which we take into account in the model. State V represents a substring (sequence fragment) in which the ends are definitely base-paired. States W and W B represent a substring in which the ends are either paired or unpaired.
Description of the probabilistic RNA model for local gapped alignments. The double-circled states O L , O R , O J (defined as in Figure 5) represent composite states responsible for possible independently aligned emissions within the RNA model.
To extend these more or less standard RNA folding algorithm conventions from a single sequence to an aligned pair of sequences, let us introduce some vectorial notation. In this notation

stands for the corresponding positions i in sequence X and i' in sequence Y. Similarly  stands for the pair of nucleotides in positions i and i' of sequences X and Y respectively. With this notation, we also define
stands for the pair of nucleotides in positions i and i' of sequences X and Y respectively. With this notation, we also define  and
and  . We are going to assume that for two aligned columns
. We are going to assume that for two aligned columns  and
and  , x
i
is base-paired to x
j
if and only if xi' is base-paired to yj' which is a reasonable assumption if we are trying to find commonly occurring secondary structures within an alignment of two sequences.
, x
i
is base-paired to x
j
if and only if xi' is base-paired to yj' which is a reasonable assumption if we are trying to find commonly occurring secondary structures within an alignment of two sequences.
W acts as the starting state. W and W B are essentially equivalent, but W B is used exclusively for starting multiloops. The production rules for W are (for W B , replace W by W B everywhere in the recursion),


The vector

provides us with a compact notation to represent the three possible situations in which one nucleotide is emitted in at least one of the two sequences in the alignment. The components ex and ey take values 1 or 0 with at least one of the two being different from zero. If ex = 0 or ey = 0 we place a gap in the corresponding position in the alignment.
The symbol V represents the paired state, that is, the state we are in after emitting one pair in each sequence. The recursion for state V is,

Here the first transition corresponds to hairpin loops, and is equivalent to function FH(i, j) in [23]; the second transition corresponds to stems, bulges, and internal loops, and is equivalent to function FL(i, j, k, l) in [23]; the last transition corresponds to multiloops, that is, loops closed by more than two hydrogen bonds. The length of the alignments generated for those hairpin loops and bulges and internal loops is variable and depends on the number of gaps introduced. The only condition is that all nucleotides in that segment have to be used – for instance, and
and  for the hairpin loops.
for the hairpin loops.
The full description of the algorithms associated to this grammar is given in the Additional file. The algorithms requires two kind of emission probabilities,
-
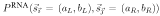
, the concurrent emission of two paired nucleotide in both sequences, already introduced in equation (6).
-
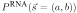
, the concurrent emission of one unpaired nucleotide in both sequences, which are taken as the mutation probabilities in equation (4).
Both types of emission probabilities have been extended to also emit gaps. For any position

, we also introduce a penalty for "mutating" to a gap, and another one for "pairing" to a gap. This is a linear gap cost, and is more convenient than implementing the additional extra states that an affine gap cost would require.
The vectorial notation becomes particularly important if we realigned the input sequences to the RNA model. In this paper, though, we will only be working with a special case where we hold an input (BLASTN) pairwise alignment fixed and simply score it with the RNA model; in this case, for any given vector

.
Transition Probabilities
In all three models, one of the prices we pay for introducing realistic flexibility is that we have introduced a number of transition probability parameters, in addition to the emission probabilities presented in the ungapped case (Section 2.1). Now we have to determine the transition probabilities of the different models. Again, we want the models tuned to the same level of "gappiness", else alignments may be artifactually classified based on how gappy they are. Whereas we were able to construct divergence-matched emission probabilities for the three models in a somewhat justified fashion, we have no guiding theory for constructing divergence-matched transition probabilities.
Instead, we have estimated all new transition probabilities by hand. The number of additional parameters in the most complete models is 8 for the OTH model, and 20 for the COD model and RNA models. These parameters have been optimized by studying the algorithm's discrimination ability on model generated data and random sequence alignments. More details on the type of simulated data used to set the transition probabilities of the models is given in Section 3.1. This approach to estimating the transition parameters of the models is very arbitrary, but we do not currently see a plausible alternative.
The RNA model also has additional SCFG-related probability parameters to take into account length distributions of hairpin loops, bulges, and internal loops. Those parameters have been determined from a training set of aligned tRNAs and ribosomal RNAs as described previously [12, 26, 27].
Alignment and scoring algorithms
We are given a pairwise sequence alignment

, composed of L aligned columns. We will hold the input alignment fixed. Thus, globally aligning and scoring the alignment with each of the three models could be done by straightforward extensions of standard HMM Viterbi and/or Forward algorithms and SCFG CYK and/or Inside algorithms. The OTH and COD pair-HMM alignment algorithm would cost O(L) in storage and time; the RNA pair-SCFG algorithm would cost O(L2) in storage and O(L3) in time [12, 24].
We are interested, however, in a combination of the standard algorithms. Consider the RNA model. Recall that we need to obtain P( |RNA) by a summation over all possible structures, which will require the Inside algorithm rather than the CYK algorithm (which, like Viterbi for HMMs, recovers the maximum likelihood parse, i.e. structure). But we will also be interested in obtaining a maximum likelihood location of a predicted RNA within the input alignment – that is, we would like to identify the maximum likelihood position of the starting and ending nucleotides that are aligned to states in the core of the RNA model, as opposed to flanking states that are accounting for flanking nonconserved (IID) and conserved but non-RNA (OTH) nucleotides in the input alignment. This would require the CYK (maximum likelihood parse) algorithm for the RNA model, outside of the core RNA pair-SCFG state.
|RNA) by a summation over all possible structures, which will require the Inside algorithm rather than the CYK algorithm (which, like Viterbi for HMMs, recovers the maximum likelihood parse, i.e. structure). But we will also be interested in obtaining a maximum likelihood location of a predicted RNA within the input alignment – that is, we would like to identify the maximum likelihood position of the starting and ending nucleotides that are aligned to states in the core of the RNA model, as opposed to flanking states that are accounting for flanking nonconserved (IID) and conserved but non-RNA (OTH) nucleotides in the input alignment. This would require the CYK (maximum likelihood parse) algorithm for the RNA model, outside of the core RNA pair-SCFG state.
To combine the desired features of the two algorithms, we use a trick introduced by Stormo and Haussler [28], perhaps most widely known for its application in the "semi-Markov model" of the (protein) genefinding program GENSCAN [29]. The basic idea is that we start with a model organized into "meta-states" (such as the O B , O E , O J , and RNA states of the RNA model in Figure 7). Each meta-state contains its own (possibly complex and arbitrary) model of a feature (such as the pair-SCFG represented by the RNA state). The meta-states are connected to each other by transition probabilities as in an HMM. To parse and score a sequence, "feature scores" are first precomputed for the score of all possible subsequences i..j being generated by each meta-state; then a dynamic programming algorithm is used to assemble a maximum likelihood parse of the sequence into a series of component features. Thus, we can (for instance) use a pair-SCFG Inside algorithm to precompute scores W ij for the core RNA metastate of the RNA model generating the part of the alignment from i..j summed over all possible structures, then use the Stormo/Haussler parsing algorithm to determine the optimal i..j segment that should be assigned as structural RNA, versus assigning flanking sequence to the O B , O E or O J meta-states describing non-RNA conserved residues and nonconserved residues.
Stormo/Haussler parsing algorithms add one order of complexity both in storage and time to the underlying dynamic programming problem to which they are applied. The Forward algorithm for scoring a pair-HMM against a fixed pairwise alignment is O(L), but since HMM dynamic programming algorithms work by iteratively calculating scores of prefixes 1..j of increasing length, whereas we need scores of subsections i..j, we have to run the algorithm L times, once from each possible start point i, making the feature scoring phase O(L2) in storage and time for both the OTH and the COD pair-HMM. (The COD pair-HMM would be O(L3) in memory, but the actual implementation uses a simplified O(L) version for the OTH meta-states included in the COD model that keeps the whole COD parsing algorithm O(L2).) The Inside algorithm for scoring a pair-SCFG against a fixed pairwise alignment is O(L2) space and O(L3) time, and conveniently yields the matrix of scores we need for all subsections i..j. Therefore the computational complexity of our complete algorithm is dominated by the Inside algorithm for scoring the core RNA state of the RNA model. (See Additional file for more details.)
In principle, we could forget about the input pairwise alignment, and allow our three models to optimally realign the input sequences. This would be desirable; it is dangerous, for example, to rely on the external sequence alignment program (e.g. BLASTN) to produce a correct secondary structural alignment of two homologous RNAs, whereas the RNA pair-SCFG, which models base-pairing correlation, would potentially produce better structural alignments. However, such an algorithm would be expensive: for two input sequences of length m and n respectively, scoring the RNA pair-SCFG would cost O(m2n2) in storage and O(m3n3) in time. (See the Additional file for a detailed description of all the different algorithms, and their complexity.) Since this realignment approach is prohibitive, we rely on an assumption that the external pairwise alignment algorithm will produce alignments that are close enough to being correct for coding regions and structural RNAs, even though the external alignment program has no notion of these constraints.
Bayesian score evaluation
Once we have calculated the probabilities that a pairwise alignment has been generated by any one of the three models, we can classify the alignment into one of three using a posterior probability calculation:

where

We assume a uniform distribution for the prior probabilities P(Model i ).
In some figures, we use a phase diagram representation of the same information in the three posterior probabilities. We plot log-odds scores of the COD and RNA models with respect to the OTH model in an (x, y) plane:

We can then separate the plane into three different regions "phases" dominated by any of the three models (for example, see Figure 8). Those three phases correspond to the conditions,
Each figure depicts 2-dimensional posterior log-odds scores for a collectionof 1,000 alignments of 200 nucleotides in length synthetically generated by the CODthe RNA and the OTH models respectively. For each figure, in blue we represent thescores of the actual alignments, while in red we represent the scores after the columnsin the alignments have been shuffled.

Points deep in one of the phases represent a higher posterior probability for a particular model, whereas points falling next to phase-transition boundaries represent situations in which the method can not clearly decide for one model or the other.
Implementation
This approach was implemented in ANSI C in a program called QRNA. The source code and the full set of probability parameters used in QRNA are freely available from http://www.genetics.wustl.edu/eddy/software/ under the terms of the GNU General Public License. QRNA has been tested on Intel/Linux and Silicon Graphics IRIX platforms.
The input alignment is given in a modified (aligned) FASTA file format. For instance the following file contains the two homologous nematode sequences shown in the BLASTN alignment in Figure 3:
>F08G2
CATTTCATAGTGTCACACGCGCACCCATGAGTTGTCGGCACAC-CACTCCCCACTACCCCTACCCTCTCCTCCATTCAGTATCGCTTCTTCGGCTTATTAGCTAAGATCAAAGTGTAGTATCTGTTCTTATCGTATTAACCTACGGTATACACTCGAATGAGTGTAATAAAGGTTATATGATTTTTGGAACCTAGGGAAGACTCGGGGCTTGCTCCGACTTCCCAAGGGTCGTCCTGGCGTTGCACTGCTGCCGGGCTCGGCCCAGTCCCCGAGGGGACAA
>G42J05
CATTCCATAGTGGCCGACGCGAGCCCGGTTTTTGTCGGTACATGCGCGCACC-CTACCCCCCGCGCCTCGTTCTCACCGCATCGCTTCTTCGGCTTATTAGCTAAGATCAAAGTGTAGTATCTGTTCTTATCGTATTAACCTACGGTATGCACTCGAATGAGTGTAATAAAGGTTATATGATTTTTGGAACCTAGGAAAGACTCGGGGCTTGCTCCGACTTTCCAAGGGTCGTCCCGGCGTTGCACTGCTGCCGGGCTCGGCCCAGTCCCTGTGGGGACAA
Note the gap characters preserving the pairwise alignment. (In many cases, there would be more gap characters than in this particular example.) Multiple pairs of sequences can be added to a single fasta file, and will be scored sequentially, one pair at a time. Typing the following command line:
qrna fastafile
we obtain the output in the following form:
>F08G2 (281)
>G42J05 (281)
... [some irrelevant output not shown]...
winner = RNA
OTH = 152.817 COD = 129.240 RNA = 182.522
logoddspostOTH = 0.000 logoddspostCOD = -23.577 logoddspostRNA = 29.705
The line winner = RNA indicates that the 281 nt alignment has been classified as a structural RNA. The next three numbers correspond to the P(

|Model) in log-odds scores. The two non-null numbers in the second row ("logoddspostCOD"and "logoddspostRNA") correspond to the 2-D phase diagram scores described previously. For this alignment, the RNA model is favored over COD and OTH by 29.7 bits.
A scanning version of the algorithms is also implemented. In this scanning mode a partial segment of the alignment – a window of user-determined fixed length – is scored. The window slides across the alignment and each window is scored independently from the others. This option is useful when the input alignment is long, or one expects different types of functionalities within a given alignment. This is the mode of the program that we use for whole genome analysis.
Scoring a window of 200 nts takes about 14 CPU-seconds and 8 MB of memory on 225 Mhz MIPS R10K processor of a Silicon Graphics Origin2000. Scoring an alignment of 2 Kbases in windows of 200 nts and moving 50 nts at a time takes about 9 minutes. Scoring the alignments generated between the intergenic regions of E. coli and S. typhi (12, 000 alignments with average length of about 100 nt) took about 9 CPU-hours.
Results
Tests on simulated data
Because all three models are fully probabilistic, we can use them in a generative mode to sample synthetic pairwise alignments. These simulations allow us to assess the sensitivity and specificity of the approach on idealized data, to get a sense of the best that the algorithm can do. We generated 1, 000 pairwise alignments of 200 nucleotides in length from each of the three models. Each of these 3, 000 alignments was then scored and classified by the program. Results are shown in Figure 8, showing that simulated alignments are almost always classified correctly.
We wanted to test that the classification is based on the pattern of mutation in the alignments, not on a spurious artifact of differing base composition, sequence identity, or gap frequency. To do this, we randomly shuffled each alignment by columns – preserving the sequence identity in the alignment, while destroying any correlations in the pattern of mutation. Figure 8 shows that shuffled alignments are classified in the OTH phase, as expected.
These simulation experiments were iterated during the development of the approach. They were important guide in setting the ad hoc transition probabilities in each model.
We used these simulation results from RNA-generated and shuffled data to set crude but reasonable score thresholds for classification of alignments as RNA. A threshold of 1.4 bits for the RNA posterior log-odds scores would determine a minimum error rate area with a frequency of 0.023 false positives and 0.081 false negatives. In whole-genome scans, we want to push the rate of false positives down, even at the expense of increasing the number of false negatives. To reduce the false positive frequency to 0.005, we would need a cutoff of 5.8 bits, which increases the false negative frequency to 0.14. We set a cutoff of 5 bits for the remainder of the results in this paper. These error rates are probably somewhat pessimistic. Figure 8 shows that the rate of false positive RNA classifications of COD or OTH-generated data is lower (about 0.001) at the 5-bit cutoff that we set based on finding false positives in shuffled RNA-generated alignments.
Tests on simulated genomes
To get a better idea of the false positive rate in whole genome screens, where the background is dominated by sequences other than RNAs, we used the COD and OTH models to simulate two aligned complete "pseudobacterial" genomes with no structural RNA genes present. The aligned pseudo-genomes have the following characteristics [30, 31]: ~2 megabases in total length, with coding regions generated from the COD model with length distributions distributed normally around a mean length of ~900 nucleotides, and intergenic regions generated using the OTH model with length distributions distributed normally around the mean length of ~100 nucleotides (thus, an overall coding density of ~90%).
Because the parameters of the models are ultimately dependent upon the BLOSUM62 amino acid scoring matrix, the average percent identity of the aligned genomes was only 41% in "coding" regions and 36% in "intergenic" regions. This is a weakness in the simulation, because in a real genome screen, we would be looking at alignments in the 65–85% nucleotide identity range, as we discuss later in the paper.
The parameters also gave a simulated pair of genomes with an overall GC content of 47.25%. From previous experience [12], we expected that genomic sequences with high GC content might tend to be misclassified as RNAs. We therefore devised a crude way of modifying the parameters of the models to correspond to different base compositions, by expressing various joint probabilities instead as a function of conditional probabilities, i.e. for two aligned codons c, c':
P (c, c') = P(c|c')P(c'), (16)
where the P(c') are codon frequencies obtained by marginalization of the joint probabilities, and the information about the mutation rate is in the conditional probabilities P(c|c'). We then can modify overall frequencies to a different set  while keeping the same conditional probabilities. The joint probabilities are then recalculated as:
while keeping the same conditional probabilities. The joint probabilities are then recalculated as:

where

is obtained as the product of the single nucleotide frequencies for the new GC composition. This approximation for the codon probabilities could be refined to better reflect the actual codon bias of the genome.
This correction of probabilities can be performed for both codon-codon probabilities and independent mutation probabilities. Using this modification to the COD and OTH models, we generated two more pairs of genomes which had overall GC contents of 57.7% and 38.9%. We then ran QRNA using its default parameters (i.e. uncorrected for GC composition) across these three aligned simulated genomes in scanning mode, using a window 200 nucleotides wide, moving 50 nucleotides at a time, and counted the number of times a window was called RNA with a score of ≥ 5 bits. All such windows are false positives, because the simulated genomes have no RNA component.
The observed false positive numbers for the 2 Mb low-GC, average-GC, and high-GC simulated genomes were 8, 14, and 21 respectively, or about 4–10 per megabase of pairwise alignment analyzed. This indicates that specificity degrades with higher GC compositions. We reanalyzed the high-GC genome using the high-GC parameter set that generated it (i.e. parameters corrected for GC composition), and saw one false positive. This indicates that setting the parameters of the three models to be appropriate for the GC composition of the input alignment should improve the effectiveness of the approach; however, our current method for doing this may be too crude.
Tests on known RNAs
To test the sensitivity and specificity of our method on real RNAs, we analyzed pairwise alignments taken from a multiple alignment of 63 eukaryotic SRP-RNAs [32] (also known as 7SL RNA), and a multiple alignment of 51 eukaryotic RNaseP RNAs [33]. These RNA genes were chosen because they are independent from the set of tRNAs and rRNAs used to train the RNA model.
We did two different types of experiments. In the first, we used the pairwise alignments as given in the curated multiple sequence alignment. These pairwise alignments are an ideal case for QRNA, because they are structurally aligned. In the second set of experiments, we took each known RNA in turn and used it as a BLASTN query against the rest of the RNAs, then classified all significant alignments with QRNA. This is a more realistic scenario for QRNA; a BLASTN primary sequence alignment may be fragmentary and/or not entirely structurally correct. All alignments were scored with QRNA using default parameters.
For the first experiment, we used QRNA to score in full (i.e. not with a scanning window) the 2, 016 different structural pairwise alignments for SRP-RNAs, and the 1, 325 structural pairwise alignments for RNaseP RNAs. The manually curated RNA structural alignments have a wide range of sequence diversity that extends from 100% to 0% pairwise identity. The number of pairwise alignments that were classified as RNA with a score of > 5 bits was counted, and these counts were binned by ranges of percent identity. The fraction of alignments classified as RNA is a measure of the sensitivity of QRNA. To measure specificity, we randomly shuffled each pairwise alignment by columns, which destroys the nested RNA structure correlations but retains the percentage identity of the alignments. Shuffled alignments that are classified by QRNA as RNA are false positives. The results in Table 2 show that QRNA can detect about half of the alignments as RNAs at a wide range of percent identities; however, specificity seriously degrades for alignments over 90% identity.
In the second set of experiments, we have taken each single RNA gene in a given family (both for the SRP-RNA and the RNaseP RNA families) and used it as a BLASTN query against all genes in the same family (including itself). We used WUBLASTN (2.0MP-WashU, 12 Feb 01 version, default parameters and scoring matrix) and retained those alignments that were longer than 50 nucleotides, with an E-value of ≤ 0.01, and with an overall similarity of ≥ 65%. Of the 3, 342 possible comparisons, this produced 1, 003 alignments (586 for SRP RNAs, and 417 for RNaseP RNAs). These were then scored by QRNA to measure sensitivity, and then shuffled by columns and rescored to measure specificity. Table 3 shows that specificity follows the same trend we saw in the structural alignments, with a sharp degradation in specificity over 90% identity. Sensitivity, however, drops off steeply in the other direction; as percent identity declines, sensitivity decreases.
We also analyzed the dependency of sensitivity and specificity with the GC content of the alignments, both for structural and BLASTN-type alignments. We observe a similar trend for both types of alignments; both sensitivity and specificity reach their best values for GC contents ranging from 45% to 60%. Specificity drops faster for high GC content alignments, which is consistent with the fact that unstructured sequences with high GC content tend to produce more spurious secondary structure predictions than low GC content sequences [12].
These results show two competing forces at play. In order to be detected by QRNA, two RNA sequences must be similar enough to produce a BLASTN alignment that is reasonably correct and extensive, but they also must be dissimilar enough to show compensatory mutations in base-paired positions of the RNA secondary structure. There is therefore a "sweet spot" of percent identity in which QRNA performance is optimal. Based on these results, we choose to analyze only BLASTN pairwise alignments of between 65% and 85% nucleotide identity with QRNA. However, we do not fully understand the degradation of specificity at high percentage identities (see Discussion).
Tests on a whole genome
To test QRNA performance in a realistic whole genome screen, we used it to analyze the Escherichia coli genome by comparisons to the related genome of Salmonella typhi. We compared QRNA annotation to the curated annotation of known coding genes, ncRNAs, and intergenic regions [34]. The feature tables for version M52 of the E. coli genome includes 115 known RNA genes and 4, 290 known coding genes (ORFs). The known RNA genes include 22 rRNAs, 86 tRNAs, and 7 miscellaneous RNAs (RNase P, for example). At least 4 other known RNA genes [1, 35, 36] – csrB, oxyS, micF, and rprA – were not present in the M52 feature table.
We split the E. coli genome in three different components: 115 RNA features (a total of 40 kb, 1% of the genome), 4290 ORF features (4090 kb, 88% of the genome), and 2367 intergenic sequences of length ≥ 50 nt (500 kb, 11% of the genome). Each sequence was compared against the complete Salmonella typhi genome (Sanger Centre, unpublished genome data, http://www.sanger.ac.uk/Projects/S_typhi) using WUBLASTN, and all alignments of ≥ 50 nt with an E-value of ≤ 0.01 and a percent identity of ≥ 65% and ≤ 85% were kept. This resulted in 354 alignments to RNAs, 4, 946 alignments to ORFs, and 11, 509 alignments to intergenic regions. (The large number of alignments in intergenic regions is due to repetitive sequence families.) These alignments were then classified by QRNA in scanning mode, scoring overlapping windows of 200 nucleotides sliding 50 nucleotides at a time, and all windows with scores of ≥ 5 bits for one of the three models were annotated as RNA, COD, or OTH correspondingly.
We then looked at these data in two ways. First, how many of the known features (ncRNAs and ORFs) were detected correctly? We counted a known feature as "detected" as RNA or COD if it had one or more overlapping QRNA annotations of that type. It is possible for different parts of a long feature (especially the ORFs) to be detected with different annotations. For the 115 known ncRNAs, 33 have one or more BLASTN alignments to S. typhi in the right range, and all 33 were annotated as RNA by QRNA; none were called COD. For the 4290 known ORFs, 3181 had BLASTN alignments in the right range; 2876 were called COD, 20 were called RNA, and 184 were called both COD and RNA.
These results indicate that the sensitivity of the program is largely dependent upon the availability of appropriate comparative sequence data – only 29% of the 115 known RNAs were detected, but invariably (in this case), a failure to detect an RNA resulted from the lack of an appropriate BLASTN alignment to analyze (of 65-85% identity). Therefore sensitivity could presumably be improved by using multiple comparative genome sequences at different evolutionary distances.
A second way to look at the data is from the perspective of how many of QRNA's annotations are correct. In a postprocessing step, any overlapping windows with the same QRNA annotation were merged into a longer annotated region. A total of 148 regions are annotated in the ncRNA sequence fraction: 33 as RNA, none as COD, and 115 as OTH. 7422 regions are annotated in the ORF sequence fraction: 88 as RNA, 3397 as COD, and 3937 as OTH. 1974 regions are annotated in the intergenic sequence fraction: 351 as RNA, 61 as COD, and 1562 as OTH. Therefore QRNA annotated a total of 5614 sequence regions as OTH, of which 3937 (70%) are actually in known ORFs-this means we must interpret an OTH annotation as a catch-all "don't know" category, rather than as a conserved noncoding sequence of potential interest. QRNA annotated a total of 3458 regions as COD, of which 3397 (98%) are in known ORFs. The other 61 COD annotated regions could either be false positive calls, or could be previously undetected small coding genes.
Most interestingly, QRNA annotated a total of 472 regions of E. coli as RNA, of which only 33 (7%) are in known RNAs. It is not possible to definitively accept or reject the rest of these annotations without additional experimental data. The 88 RNA annotations that overlap known ORFs may be false positives, or may indicate cis-regulatory RNA structures that overlap coding regions. It is intriguing that a disproportionate number of QRNA's RNA annotations (74%, 351/472) were in the "intergenic" data fraction, which is only 11% of the genome – which is what we would expect to see if there were a fair number of undetected RNA features in the genome.
We examined many of these 351 regions by eye. Four of them are the four ncRNA genes (csrB, oxyS, micF, and rprA) that were not included in the M52 feature table for E. coli. Others are repetitive sequence families with conserved palindromic sequence, such as BIMEs [37]. Some correspond to known cis-regulatory RNA structures such as ρ-independent terminators (which have an RNA stem loop structure) and transcriptional attenuators. For about half of these regions, we cannot exclude the possibility that they correspond to novel RNAs, and we cannot assign a known biological role to them without additional computational or experimental evidence. A more in-depth QRNA screen of E. coli for novel ncRNAs using multiple comparative genomes from γ-proteobacteria, accompanied by experimental evidence that many of the predicted RNAs are indeed novel ncRNA genes, is presented elsewhere (E.R., R.J. Klein, T.A. Jones, and S.R.E., manuscript submitted).
Discussion
There are a number of ways in which we could improve QRNA. The three probabilistic models are calibrated to a fixed evolutionary distance. We used the BLOSUM62 substitution matrix to define the fixed evolutionary distance of our three models, and it is now quite clear that this is the wrong distance. Our models generate pairwise alignments of about 40% sequence identity. We expect on theoretical grounds that this is where the models would perform optimally on real input alignments. However, BLASTN cannot detect RNA sequences that are this diverged. Our evaluations indicated a sweet spot of 65%-85% identity for QRNA to work best in its current formulation. We suspect that we could obtain some improvement by choosing a substitution matrix corresponding to more closely related nucleotide sequences.
In principle QRNA may also be useful as a coding-region genefinder. The coding model is a fully probabilistic formalization of comparative analysis ideas used by the genefinder CRITICA [16], and by comparative exon finding approaches such as the EXOFISH vertebrate/Tetraodon comparison [38] and the human/mouse comparison in[39]. In the E. coli whole genome screen, the sensitivity and specificity of QRNA coding annotations seem quite high. We have not yet attempted to optimize the performance of QRNA for this purpose.
In terms of other QRNA improvements, it should be advantageous to make the emission and transition parameters of the models conditional on a parametric evolutionary distance. We could then optimize a maximum likelihood distance separately for each input alignment (or, marginalize over all distances, in a more Bayesian approach). This should widen the 65-85% alignment identity window that QRNA works best in – in particular, by constructing models more appropriate for nearly identical sequences, where we currently have high false positive rates.
It would be good to have more theory to guide how we produce divergence-matched transition probability parameters for the three models. We suspect our ad hoc estimation may be causing the RNA model to be favored artifactually in certain cases (less gappy alignments and longer alignments), elevating our false positive rate.
We also made a number of simplifying independence assumptions in trying to calculate QRNA's parameters all from a single chosen amino acid substitution matrix. Some of these assumptions probably reduce our performance. It would be desirable to move towards estimating parameters based on real datasets of aligned nucleotide sequences, if large enough datasets could be amassed.
We are relying on BLASTN to produce approximately correct pairwise alignments of coding regions or RNA structures, even though BLASTN is purely a position-independent primary sequence alignment program. We could instead realign the two input sequences using the pair-grammars. In principle this should increase the performance of QRNA, particularly for more dissimilar sequences. Unfortunately, alignment of two sequences to a pair-SCFG is effectively the Sankoff algorithm [40] with time and memory complexity of O(L6) and O(L4), respectively, so we will need a more clever algorithmic strategy than straightforward dynamic programming (if, indeed, dynamic programming RNA structure alignment in a four-dimensional hypercube can be called "straightforward").
Because QRNA detects conserved RNA secondary structure, it is not expected to detect ncRNAs that apparently lack significant intramolecular secondary structure, such as C/D box small nucleolar RNAs [6]. Identifying novel unstructured ncRNAs remains an entirely open problem. A pure computational approach will probably have to identify transcriptional signals – promoters, enhancers, and terminators – and this remains a difficult problem, particularly in complex genomes. Experimental screens for novel ncRNAs may prove more fruitful for unstructured ncRNAs. Expression arrays that pave the entire target genome with probes can detect novel transcripts [41], and cDNA libraries that enrich for small, nonpolyadenylated RNAs can be constructed and EST sequenced [42].
QRNA is also expected to identify cis-regulatory RNA structures in mRNAs, in addition to structured ncRNA genes. Distinguishing an ncRNA gene from a cis-regulatory RNA structure in an mRNA is nontrivial in absence of experimental evidence. This cautions against using QRNA for fully automated genome annotation and "gene counting" exercises in the way that protein genefinders like GENSCAN are used.
Instead, QRNA is best used as a computational screen for candidate ncRNA genes, after which candidate loci are further characterized both computationally and experimentally before considering them to be "genes". Both the data presented here and in a second paper detailing a careful E. coli genome screen with experimental verification of many novel ncRNA genes (E.R., R.J. Klein, T.A. Jones, and S.R.E., manuscript submitted) indicate that QRNA can be successfully used in this role. Although we have much we can do to improve its performance, we believe QRNA is the first example of a generally applicable computational genefinder for noncoding RNA genes. We expect to be able to apply QRNA – based screens for ncRNAs to a number of organisms as comparative sequence data become available – including yeast, Caenorhabditis, Drosophila, human, and several microbial systems.
Conclusions
We have described an algorithm that uses three different probabilistic models (for RNA-structure-constrained, coding-constrained, and position-independent evolution) to examine the pattern of mutations in a pairwise sequence alignment. The alignment is classified as RNA, coding, or other, according to the Bayesian posterior probability of each model. We have implemented this algorithm as a program, QRNA, which we consider to be a prototype structural ncRNA genefinding program.
References
Eddy SR: Noncoding RNA genes. Curr. Opin. Genet. Dev 1999, 9: 695–699. 10.1016/S0959-437X(99)00022-2
Erdmann VA, Barciszewska MZ, Symanski M, Hochberg A, de Groot N, Barciszewski J: The non-coding RNAs as riboregulators. Nucl. Acids Res 2001, 29: 189–193. 10.1093/nar/29.1.189
Burge CB, Karlin S: Finding the genes in genomic DNA. Curr. Opin. Struct. Biol 1998, 8: 346–354. 10.1016/S0959-440X(98)80069-9
Miyajima N, Burge CB, Saito T: Computational and experimental analysis identifies many novel human genes. Biochem. Biophys. Res. Commun 2000, 272: 801–807. 10.1006/bbrc.2000.2866
Kelley RL, Kuroda ML: Noncoding RNA genes in dosage compensation and imprinting. Cell 2000, 103: 9–12. 10.1016/S0092-8674(00)00099-4
Weinstein LB, Steitz JA: Guided tours: From precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol 1999, 11: 378–384. 10.1016/S0955-0674(99)80053-2
Bachellerie JP, Cavaille J: Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In: Modification and Editing of RNA (Edited by Grosjean H, Benne R) Washington DC, ASM Press 1998, 255–272.
Meguro M, Mitsuya K, Nomura N, Kohda M, Kashiwagi A, Nishigaki R, Yoshioka H, Nakao M, Oishi M, Oshimura M: Large-scale evaluation of imprinting status in the Prader-Willi syndrome region: An imprinted direct repeat cluster resembling small nucleolar RNA genes. Hum. Mol. Genet 2001, 10: 383–394. 10.1093/hmg/10.4.383
Lease RA, Belfort M: Riboregulation by DsrA RNA: Trans-actions for global economy. Mol. Micro 2000, 38: 667–672. 10.1046/j.1365-2958.2000.02162.x
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al.: Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408: 86–89. 10.1038/35040556
Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al.: Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 2001, 104: 195–203. 10.1016/S0092-8674(01)00205-7
Rivas E, Eddy SR: Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics 2000, 6: 583–605. 10.1093/bioinformatics/16.7.583
Le SY, Chen JH, Currey KM, Maizel JV: A program for predicting significant RNA secondary structures. Comput. Applic. Biosci 1988, 4: 153–159.
Le SY, Chen JH, Maizel JV: Efficient searches for unusual folding regions in RNA sequences. In: Structure and Methods: Human Genome Initiative and DNA Recombination (Edited by Sarma RH, Sarma MH) Adenine Press 1990, 1: 127–136.
Chen JH, Le SY, Shapiro B, Currey KM, Maizel J: A computational procedure for assessing the significance of RNA secondary structure. Comput. Applic. Biosci 1990, 6: 7–18.
Badger JH, Olsen GJ: CRITICA: Coding region identification tool invoking comparative analysis. Mol. Bio. Evol 1999, 16: 512–524.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J. Mol. Biol 1990, 215: 403–410. 10.1006/jmbi.1990.9999
Durbin R, Eddy SR, Krogh A, Mitchison GJ: Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge UK, Cambridge University Press 1998.
Altschul SF: Amino acid substitution matrices from an information theoretic perspective. J. Mol. Biol 1991, 219: 555–565.
Henikoff S, Henikoff JG: Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89: 10915–10919.
Ikemura T: Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Bio. Evol 1985, 2: 13–34.
Hopcroft JE, Ullman JD: Introduction to Automata Theory, Languages, and Computation. Addison-Wesley 1979.
Zuker M, Stiegler P: Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucl. Acids Res 1981, 9: 133–148.
Rivas E, Eddy SR: A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol 1999, 285: 2053–2068. 10.1006/jmbi.1998.2436
Rivas E, Eddy SR: The language of RNA: A formal grammar that includes pseudoknots. Bioinformatics 2000, 16: 326–333. 10.1093/bioinformatics/16.4.326
Steinberg S, Misch A, Sprinzl M: Compilation of tRNA sequences and sequences of tRNA genes. Nucl. Acids Res 1993, 21: 3011–3015.
Van de Peer Y, Van den Broeck I, De Rijk P, De Wachter R: Database on the structure of small ribosomal subunit RNA. Nucl. Acids Res 1994, 22: 3488–3494.
Stormo GD, Haussler D: Optimally parsing a sequence into different classes based on multiple types of evidence. ISMB 1994, 2: 369–375.
Burge C, Karlin S: Prediction of complete gene structures in human genomic DNA. J. Mol. Biol 1997, 268: 78–94. 10.1006/jmbi.1997.0951
Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, et al.: Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii . Science 1996, 273: 1058–1073.
Kawarabayasi Y, Sawada M, Horikawa H, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, et al.: Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res 1998, 5: 55–76.
Larsen N, Zwieb C: SRP-RNA sequence alignment and secondary structure. Nucl. Acids Res 1991, 19: 209–215.
Brown JW: The ribonuclease P database. Nucl. Acids Res 1998, 27: 314. 10.1093/nar/27.1.314
Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado Vides J, Glasner JD, Rode CK, Mayhew GF, et al.: The complete genome sequence of Escherichia coli K-12. Science 1997, 277: 1453–1462. 10.1126/science.277.5331.1453
Wassarman KM, Zhang A, Storz G: Small RNAs in Escherichia coli . Trends Microbiol 1999, 7: 37–45. 10.1016/S0966-842X(98)01379-1
Majdalani N, Chen S, Murrow J, St John K, Gottesman S: Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol 2001, 39: 1382–1394. 10.1046/j.1365-2958.2001.02329.x
Bachellier S, Clement JM, Hofnung M: Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol 1999, 150: 627–639. 10.1016/S0923-2508(99)00128-X
Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fischer C, Fizames C, Wincker P, Brottier P, Quetier F, et al.: Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet 2000, 25: 235–238. 10.1038/76118
Batzoglou S, Pachter L, Mesirov JP, Berger B, Lander ES: Human and mouse gene structure: Comparative analysis and application to exon prediction. Genome Res 2000, 10: 950–958. 10.1101/gr.10.7.950
Sankoff D: Simultaneous solution of the RNA folding, alignment, and proto-sequence problems. SIAM J. Appl. Math 1985, 45: 810–825.
Selinger DW, Cheung KJ, Mei R, Johansson EM, Richmond CS, Blattner FR, Lockhart DJ, Church GM: RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nature Biotech 2000, 18: 1262–1268. 10.1038/82367
Huttenhofer A, Kiefmann M, Meier Ewert S, OBrien J, Lehrach H, Bachellerie JP, Brosius J: RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 2001, 20: 2943–2953. 10.1093/emboj/20.11.2943
Thomas J, Lea K, Zucker Aprison E, Blumenthal T: The spliceosomal snRNAs of Caenorhabditis elegans . Nucl. Acids Res 1990, 18: 2633–2642.
Acknowledgments
We thank Jan Gorodkin for pointing to us some multiple alignments for RNA genes, and Ian Holmes for critical reading of the manuscript. The work of E.R. was partially supported by a postdoctoral grant from the Sloan Foundation. We thank the Howard Hughes Medical Institute and the NIH National Human Genome Research Institute for other funding, and Silicon Graphics, Sun Microsystems, Compaq, Paracel, Hewlett-Packard, IBM, and Intel Corporation for continued hardware and engineering support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
12859_2001_9_MOESM1_ESM.pdf
Additional file: • Description of data: In this additional file we provide a detailed description of the algorithms involved in implementing the three probabilistic models components of our comparative method QRNA. We give the most general description of the scoring/parsing algorithms. We also indicate how to obtain some simplifications that are part of the software implementation. (PDF 116 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Rivas, E., Eddy, S.R. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics 2, 8 (2001). https://doi.org/10.1186/1471-2105-2-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2105-2-8