- Methodology article
- Open access
- Published:
Pairwise alignment of nucleotide sequences using maximal exact matches
BMC Bioinformatics volume 20, Article number: 261 (2019)
Abstract
Background
Pairwise alignment of short DNA sequences with affine-gap scoring is a common processing step performed in a range of bioinformatics analyses. Dynamic programming (i.e. Smith-Waterman algorithm) is widely used for this purpose. Despite using data level parallelisation, pairwise alignment consumes much time. There are faster alignment algorithms but they suffer from the lack of accuracy.
Results
In this paper, we present MEM-Align, a fast semi-global alignment algorithm for short DNA sequences that allows for affine-gap scoring and exploit sequence similarity. In contrast to traditional alignment method (such as Smith-Waterman) where individual symbols are aligned, MEM-Align extracts Maximal Exact Matches (MEMs) using a bit-level parallel method and then looks for a subset of MEMs that forms the alignment using a novel dynamic programming method. MEM-Align tries to mimic alignment produced by Smith-Waterman. As a result, for 99.9% of input sequence pair, the computed alignment score is identical to the alignment score computed by Smith-Waterman. Yet MEM-Align is up to 14.5 times faster than the Smith-Waterman algorithm. Fast run-time is achieved by: (a) using a bit-level parallel method to extract MEMs; (b) processing MEMs rather than individual symbols; and, (c) applying heuristics.
Conclusions
MEM-Align is a potential candidate to replace other pairwise alignment algorithms used in processes such as DNA read-mapping and Variant-Calling.
Background
Biological sequence alignment [1] is about finding similarities and differences between sequences. The term alignment covers a broad range of different processes. Seed-and-extend alignment method is a popular technique for aligning reads to the reference-genome. This technique is used in DNA read-mappers such as BWA [2, 3] and Bowtie [4, 5]. In the seed-and-extend technique, small subsequences of a read (called seeds) are searched in the reference-genome to find candidate regions. Once a rough alignment is identified (seeding-step), the read is typically aligned to all candidate regions using a dynamic programming algorithm (extending-step)
Seed-and-extend method is also used in similarity search tools such as BLAST [6], BLAT [7] and MUMmer [8] as well as PatternHunter [9] and PatternHunter II [10]. In order to search for the seed in the reference-genome, some methods such as BWA and Bowtie use a suffix-tree-based structure called FM-Index [11] while others such as BLAST and SNAP [12] use a hash-table index of fixed size k-mers (subsequences of length k).
The seeding-step varies from program to program. For example, BWA-MEM [3] looks for Maximal Exact Matches (MEMs) and MUMmer looks for Maximal Unique Matches (MUMs). GEM [13] limits the seed size to have at least n+1 non-overlaping seed to find all alignments with up to n errors. Another technique is to use spaced-seeding [14] that is used in PatternHunter and PatternHunter II.
The focus of this paper is the extending-step and not the seeding-step. Typically, in the extending-step, dynamic programming is used. Dynamic programming alignment could produce global [15], local [16] or semi-global [1] alignment. Such an extending-step could also implement different scoring systems such as edit-distance [17] scoring or affine-gap [18] scoring.
Most DNA read-mappers [2–5, 19] use a derivative of the Smith-Waterman algorithm which uses affine-gap scoring to find a semi-global alignment. Such implementations of the Smith-Waterman algorithm can be found in [20–23]. These implementations exploit data-level parallelisation (SIMD) instructions of Intel processors (SSE) to further speed up the alignment process.
Not all DNA read-mappers use the above extending-step. For example, GEM and SNAP use modified version of Gene Myers [24] and Ukkonen [25] algorithms respectively for their extending-step. Both Gene Myers and Ukkonen algorithms are edit-distance based dynamic programming alignment. There are other alignment algorithms which do not use the conventional dynamic programming technique. For example, PatternHunter uses its own custom-made extending-step. Another example is a greedy approach for aligning DNA sequences introduced in [26]. The Smith-Waterman algorithm is also accelerated on Graphic Processing Units (GPUs) [27] and Field Programmable Gate Arrays (FPGAs) [28, 29]. However, the scope of this paper is the use in conventional computing platform (CPUs), though they can be extended for use in FPGAs.
In this paper, we present a dynamic programming alignment algorithm called DP-MEM to be used in the extending-step of the DNA read-mapper tools. Our algorithm produces a semi-global alignment in which the first few bases or the last few bases can be excluded (clipped) from the alignments (not affecting alignment score). Also, the proposed algorithm implements the affine-gap scoring model. Unlike previous dynamic programming alignment algorithms which work on individual bases of the sequences (see Fig. 1, DP-MEM works on the MEMs which exist between a pair of short sequences.
DP-MEM requires all MEMs to be extracted from a given pair of sequences. To reduce the overhead of extracting MEMs, we introduce a bitwise parallel method to quickly extract all MEMs which exist between a pair of short sequences. DP-MEM requires MEMs to be sorted. Thus, a linear counting sort algorithm is used to speed up the sorting of extracted MEMs (Additional file 1: Section I). DP-MEM along with our proposed MEM extraction method and several heuristic optimisations introduced in this paper form a fast and near-accurate alignment method called MEM-Align.
We motivate the work in this paper with the following simplified example. Consider aligning the following sentences:
-
my dog is friendly whenever playing in the park
-
all dogs are friends when playing in parks
It is clear that matching words such as “dog”, “friend” and “park” rather than individual letters such as “m”, “y” and “d” would speed up the alignment process. MEM-Align uses MEMs (similar to matching words in sentences) to align sequences. MEM-Align is suitable for aligning similar sequences (i.e. a read and its candidate region in the reference-genome) where the number of existing MEMs are significantly smaller than the number of bases in the sequences.
Figure 2 identifies possible applications of MEM-Align. In addition to DNA read-mapping software, MEM-Align can be used in Variant-Callers such as GAKT HaplotypeCaller [30] and Platypus [31] to align haplotypes to reference-genome and reads to haplotypes. MEM-Align can be used for all applications where a pair of short and similar DNA sequences are aligned.
To show the efficacy of our work, we compare the speed and accuracy results to an accelerated implementation of the Smith-Waterman algorithm as well as algorithms proposed by Gene Myers and Ukkonen.
Note that, Maximal Exact Matches has been used in the seeding-step [3]. Although MEM-Align uses MEMs, it is not a seeding method and should not be compared with the entire seed-and-extend alignment. MEM-Align should be considered a replacement for algorithms used in the extending-step.
Method
Approach
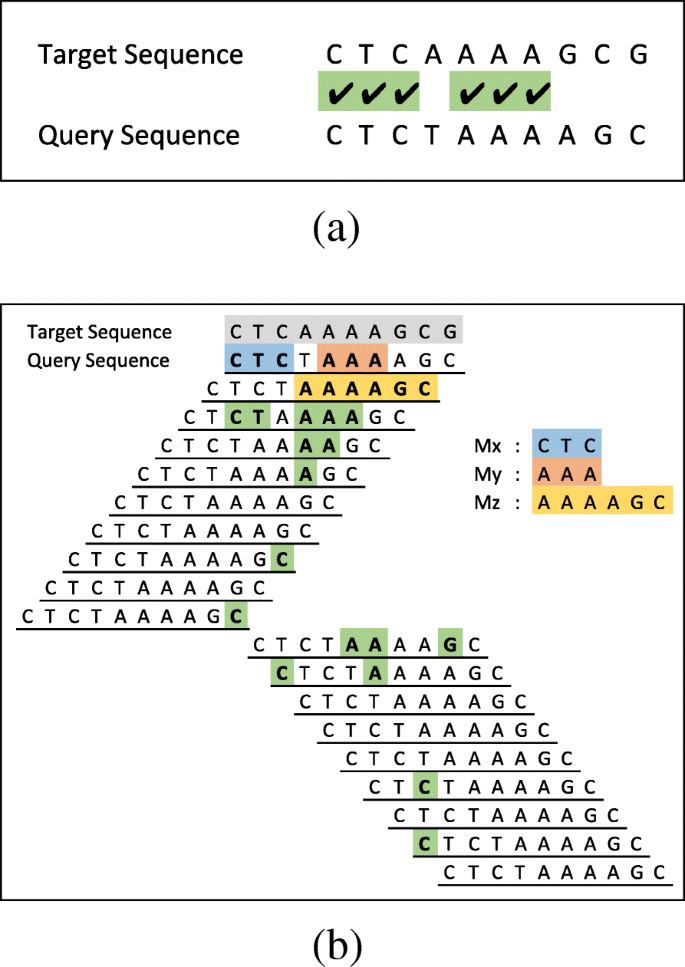
In our proposed algorithm, the first step towards aligning sequences is to extract MEMs between sequences by directly comparing them. Figure 3a is an example which compares a target and a query sequence where CTC and AAA are two MEMs identified by the comparison. Each group of continuous identical symbols in the comparison, result in a MEM even if it is composed of only a single matching symbol. In order to extract all MEMs between the sequences, the query sequence must be shifted all the way to the right and to the left one symbol at a time (see Fig. 3b). After each shift, the comparison step must be repeated to identify new MEMs. For example, the third line in Fig. 3b represents the case where the query sequence is shifted to the right one symbol and is compared with the target sequence. The result of the comparison identifies AAAAGC as a new MEM. All other MEMs extracted by shift and compare operations are also highlighted in Fig. 3b. Three of the MEMs (Mx,My and Mz) are highlighted with different colours to be used for later explanation.
MEM extraction using shift and compare operations. a Identify MEMs by direct comparison of sequences. b The query is shifted to left until last symbol in query sequence is aligned to the first symbol in target sequence. Then the query sequence is shifted to right until the first symbol in query sequence is aligned to the last symbol of target sequence. After each shift the overlapping part of the query and target sequences are compared to identify new MEMs. Three of MEMs (Mx,My and Mz) are highlighted with different colours to be used for later explanation
In the affine-gap scoring model, the alignment score AS is computed using Eq. 1 where Nm is number of matches each receiving a match score of Rm,Nx is number of mismatches each receiving a mismatch penalty of Px,No is number of gap openings each receiving a gap open penalty of Po and Ng is total length of all gaps, each gap receiving a gap extension penalty of Pg. There would be a gap opening for each group of continuous gap. For example, if there are two gaps in the alignment, where the length of the first gap is three and the length of the second gap is four, then there are two gap openings (No=2) and the total length of the gap is seven (Ng=3+4=7).
Given the list of all MEMs, the alignment can be computed using partial alignments. For example, consider MEMs Mx, My and Mz in Fig. 3b. The partial alignments made by taking different combinations of Mx, My and Mz along with the number of matches, mismatches and gaps, as well as the resulting alignment scores are shown in Fig. 4. The alignment that only includes Mx and Mz results in the highest alignment score. Note that, My and Mz overlap each other and when both are considered in the same alignment the overlap is excluded from Mz. Considering all MEMs in Fig. 3b results in many more combinations where none of them achieves a higher score.
Examining all possible combination of MEMs would be exhaustive. In “Alignment algorithm” section we describe a novel dynamic programming algorithm DP-MEM that efficiently finds the best combination without considering all cases. DP-MEM needs to know which parts of the sequences match but not the actual symbols in the sequences. The input to DP-MEM is the positioning of MEMs in the target and in the query sequences which are obtained during the MEM extraction process described in “MEM extraction” section. How MEMs are represented with their positions and how the number of matches, mismatches and gaps are computed when MEMs are combined in an alignment are explained in the remainder of this section. Figure 5 is another example alignment with six MEMs (M1 to M6) that form the alignment between target sequence T and query sequence Q. For simplicity there is no overlap between MEMs in this example. Each MEM Mi is represented as a triplet of integer numbers: the starting positions in T and in Q (STi and SQi respectively) and its length (Li). The ending positions in T and in Q can be computed later (Φ2E of Algorithm 2). Table 1 lists the length and the positioning of M1 to M6 in T and in Q.
The number of mismatches and gaps between adjacent MEMs Mi and Mj (Mi is on the left of Mj) is computed from their positioning in sequences. First, the distance between Mi and Mj in T and in Q denoted by \({LT}_{i}^{j}\) and \({LQ}_{i}^{j}\) respectively are computed (Φ2F of Algorithm 2) then the number of mismatches between Mi and Mj, denoted by \(N_{x}^{i,j}\), is the minimum of \({LT}_{i}^{j}\) and \({LQ}_{i}^{j}\). The length of the gap between Mi and Mj, denoted by \(N_{g}^{i,j}\), is equal to the absolute value of \({LT}_{i}^{j}-{LQ}_{i}^{j}\) (Φ2G of Algorithm 2). A negative value of \({LT}_{i}^{j}-{LQ}_{i}^{j}\) indicates an insertion (there are more symbols in the query sequence) and a positive value indicates a deletion (there are more symbols in the target sequence).
For example, consider M2 and M3 in Fig. 5 where there are three symbols between them in T (\({LT}_{2}^{3}=3\)) and only two symbols between them in Q\(\left ({{LQ}_{2}^{3}=2}\right)\). This situation indicates two mismatches and a gap (deletion). Table 2 elaborates how the number of mismatches and gaps are computed for the example alignment in Fig. 5.
In case there are both mismatches and gaps between Mi and Mj, all gaps are considered continuous to reduce the gap open penalty (only one gap open penalty is applied for a continues gap). Thus, for all adjacent MEMs that have gaps between them, only one gap open penalty is applied. The placement of mismatches and the only continuous gap is not important, as it would not affect the alignment score. We assume that the mismatch penalty is constant (this is usual for DNA sequences).
If there is an overlap between Mi and Mj either in the target sequence or in the query sequence, the overlap should be excluded from Mj. The length of overlap \({MO}_{i}^{j}\) is the maximum of the length of overlap in target and in the query (Φ2B of Algorithm 2). To exclude overlap, \({MO}_{i}^{j}\) should be added to STj and SQj and subtracted from Lj (Φ2D of Algorithm 2).
MEM extraction
There are methods [3, 32] to extract maximal exact matches between lengthy sequences such as an entire genome. However, these methods are based on preprocessing and indexing of one or both sequences which is a time-consuming operation. For example, in DNA read aligner, the reference-genome is indexed once, and the same index is used each time a new read is aligned. We are looking for a quick algorithm to identify MEMs between relatively short sequences that change for each alignment. A brute force method for this problem (Additional file 1: Section II) is slow and inefficient (with the complexity of O(n3)). We propose a fast bit-level parallel method to speed up the MEM extraction process. Our MEM extraction method is based on the shift and compare operations shown in Fig. 3b. The first step is to represent sequences with bit-vectors, where A, C, T, and G are encoded as 00, 01, 10, and 11, respectively (Additional file 1: Section III). Figure 6 illustrates an example sequence pair, along with corresponding bit-vector representations. In a commodity computer, the machine word is usually 64 bits which can accommodate 32 nucleotide symbols. Since a sequence is usually larger than 32 symbols, the corresponding bit-vector is stored in multiple machine words. Each operation on bit-vectors of sequences of size n symbols acts on \(\lceil \frac {n}{32} \rceil \) machine words.
With bit-vectors representation of sequences, shifting a sequence by one symbol is the same as shifting the bit-vector by two bits, and comparing sequences can be done with XOR instruction (32 symbols at a time). In the XOR output (X), 00 means that symbols are matched, and a sequence of 00s shows a MEM. A set of shift and bitwise operations as shown in Algorithm 1 computes X and subsequently the edge bit-vector (E) in which the start and the end of each MEM are highlighted with set bits (bits with a value of one). Figure 6 shows the X and the E bit-vectors with highlighted MEMs. The positioning of MEMs in sequences are then computed from the edge bit-vector (Additional file 1: Section IV).

Alignment algorithm
In “Approach” section, we show that by considering different combinations of MEMs and computing the alignment score for the corresponding alignment, one can identify the combination of MEMs that results in the maximum alignment score. However, examining all possible combination of MEMs is a naive solution. A more systematic way of finding the alignment efficiently is to use dynamic programming.
Dynamic programming is the method of approaching the solution to a problem by defining and solving smaller subproblems. Solutions for subproblems are used to solve a bigger problem at each step. The process is repeated until all subproblems are solved. Eventually, the solution to one of the subproblems would be the solution to the initial problem. When all subproblems are solved a backtracking process identifies a series of solutions that contribute to the final solution. In dynamic programming, there should be an ordering of the input data along which the recursion procedure proceeds.
We sort all MEMs according to the position of their end in query sequence (EQ). MEMs which end in the same position are ordered in an arbitrary way. The jth subproblem is to find the alignment of subsequences of T and Q which end at jth MEM Mj (T[1...ETj] and Q[1...EQj] respectively). We will show that this ordering of MEM is sufficient to support the correct recursion.
In the sorted list of MEMs, EQi=EQj indicates that one of Mi or Mj fully overlaps the other MEM in the query sequence. Since in Φ2B of Algorithm 2 the overlap region is excluded, Mi and Mj cannot be in the same alignment. Thus ith and jth subproblems are solved independently from each other and the order of i and j in the sorted list could be arbitrary. If EQk>EQj (k>j in the sorted list), Mk could not be a part of the alignment that ends in Mj. Thus the jth subproblems can be solved independently from the solution to the kth subproblem. Note that it is also possible to sort MEMs based on their ending position in the target sequence (ET) using a similar justification.
Our proposed dynamic programming algorithm (DP-MEM) is elaborated in Algorithm 2. For the example MEMs extracted in Fig. 3b, the dynamic programming table and intermediate value computed in the algorithm are shown in Figs. 7 and 8 respectively. The input to DP-MEM is the list of MEMs where each MEM (Mj) is a triplet of integers [Lj,SQj,STj]. The second input n is the number of MEMs in the list. The output S is the alignment score for the sequences. The algorithm prints out the indices of all MEMs that forms the alignment where the first and the last printed numbers are the indices of the rightmost and the leftmost MEMs in the alignment respectively. All steps of Algorithm 2 are commented in the following:
Dynamic programming table used in Algorithm 2 to process extracted MEMs in Fig. 3b. Cell i and j represent the value of \(S_{i}^{j}\). Empty cells are not evaluated in Φ2. Evaluation of cells with cross mark are skipped in Φ2A. Initial Value of Sj is computed in Φ1. Final value of Sj and its source (what maximises Sj) are highlighted for each row. The highest Sj (S13) is the alignment score. M13 is the last MEM in the alignment and the MEM before that is MW[13]=M3. Since W[3]=−1,M3 is the first MEM in the alignment. The scoring system for this alignment is Rm=2,Px=3,Po=4 and Pe=1
Intermediate values to compute \(S_{i}^{j}\) in Fig. 7. Note that Sij in this figure refers to \(S_{i}^{j}\)
-
Φ1: Scoring each MEM for all of its matching symbols. Note that there are Lj matching symbols in Mj. Sj represents the highest alignment score for the alignment ending at Mj. Initialising Sj in this step is similar to computing the partial alignment score when only Mj is included in the alignment. W[ j] is used for backtracking. The value of -1 indicates that the current Sj is obtained by considering Mj alone in the alignment.
-
Φ2: Computing Sj for each MEM (Mj). To compute Sj, for each MEM Mi where Mi appears before Mj in the list, the algorithm adds Mj to the alignment ending at Mi (extending previously found alignments) and looks for the extension that maximises Sj (solving a bigger subproblem using previously solved subproblems).
-
Φ2A: Skip extension when it is not possible. If ETi>ETj then Mi contains part of target sequence which is beyond the alignment ending at Mj and the extension is not possible. If EQi=EQj or ETi=ETj or SQi≥SQj or STi≥STj then one of the MEMs fully overlaps the other MEM. In this case, Mi and Mj cannot be in an alignment together.
-
Φ2B: Computing the length of the overlap between Mi and Mj. If \({MO}_{i}^{j}\) is less than or equal to zero, then no overlap exists.
-
Φ2C: Keeping a copy of Mj before excluding overlap.
-
Φ2D: If overlap exists, excluding overlap from Mj
-
Φ2E: Computing ending position of Mj in T and Q.
-
Φ2F: Computing the distance (number of symbols) between Mi and Mj in T and Q.
-
Φ2G: Computing number of mismatches and gaps between Mi and Mj.
-
Φ2H: Computing the penalty for the mismatches and gaps between Mi and Mj (\(P_{i}^{j}\)). If gap exists, only one gap open penalty is subtracted.
-
Φ2I: Computing alignment score \(\left (S_{i}^{j}\right)\) when Mj is added to the alignment ending at Mi. The score for all the matching symbols in Mj (Lj×Rm) is added to the alignment score for the alignment ending at Mi (Si). Then the penalty for the gaps and mismatches between Mi and Mj\(\left (P_{i}^{j}\right)\) is subtracted.
-
Φ2J: If extending Mj to the alignment ending in Mi results into a score \(\left (S_{i}^{j}\right)\) higher than current score for Mj (Sj) then the new score is stored in Sj. Also W[j] is set to i to keep track of the Mi that maximise the score for Mj.
-
Φ2K: Restoring the the value of Mj before exclusion so that Mj can be used in other alignment extensions.
-
Φ3: Looking for the MEM with the highest Sj. This MEM is the last MEM in the alignment (Me). The highest score (Se) is returned as S which is the highest alignment score for the given sequences. The index of the MEM that maximises Sj is stored in e to begin backtracking from Me.
-
Φ4: In the alignment, the immediate previous MEM to Me is the one that maximises the alignment score for Me. The index of such MEM is stored in W[ e]. As a result, the iteration of f←W[f] visits the index of all MEMs in the alignment. When W[f] is equal to -1, Mf is the first MEM in the alignment and the iteration is stopped.

W[j] only keeps one index for Mj. However, there might be cases that multiple MEMs maximise Sj (i.e. \(S_{j}=S_{x}^{j}=S_{y}^{j}\)). Also, there might be cases that multiple MEMs maximise the alignment score (i.e. S=Sx=Sy). Considering all these cases and backtracking through all the paths results in retrieving multiple alignments, all of which are have the same score. Reporting multiple alignment paths is not implemented in our experimental implementation, though there is no practical limitation to implementing it.
In our algorithm, we do not penalise mismatches and gaps before the first MEM and after the last MEM in the alignment. This results in a local alignment algorithm. By considering these penalties the algorithm generates a global alignment (Additional file 1: Section V).
The equation to compute \(P_{i}^{j}\) in Φ2H of Algorithm 2 assumes that there is no matching symbol between T and Q in the area between Mi and Mj (all symbols are counted as mismatches or gaps). Although this assumption is not true for all Mi, it is always true for the Mi that leads to maximum \(S_{i}^{j}\) which overrules the effect of the assumption being incorrect for other Mi. As a proof, assume there is a matching symbol in the area between Mi and Mj. The matching symbol would be a MEM (Mk). Mk is already extended to the alignment ending at Mi. Thus, when extending Mj to Mk a higher score is achieved when compared to extending Mj to Mi.
Chaining colinear seeds as discussed in [33] have been widely used in the alignment of large sequences, i.e. genome-to-genome alignment. It has been also used to identify candidate regions for a read given a set of MEMs in BWA. Chaining algorithms with scoring are similar to the dynamic programming algorithm we proposed (DP-MEM). However, there are differences that make DP-MEM suitable for pairwise alignment of short sequences. DP-MEM takes into account that all MEMs within a certain gap size are provided in the input and optimises the number of iteration in the algorithm. DP-MEM also implements a heuristic approach to compensate for the effect of short MEMs removed from the input list resulting gaps between MEMs.
Optimisation
Given sequences of length n, the algorithm to extract MEMs (provided in “MEM extraction” section) requires 2(n−1) shift and 2n−1 compare operations on bit-vectors (each act on \(\lceil \frac {n}{32} \rceil \) machine words) that result in an algorithm with complexity of O(n2) to produces edge bit-vectors for the given pair of sequences. The complexity of the function that computes positioning of MEMs from the edge bit-vector and sorts them based on EQ is yet to be added. Further, if m MEMs are extracted, Φ2 of Algorithm 2 (DP-MEM) has the complexity of O(m2). Considering the complexity of MEM extraction and DP-MEM, MEM-Align is much slower than available alignment algorithms. To speed up the process, we present several optimisations for MEM-Align which achieves faster runtime by sacrificing accuracy. On the other hand, we introduce methods to improve accuracy with a minimal performance loss.
Banded alignment
Banded alignment [34] is a known heuristic method to speed up the alignment process. This technique limits the pattern of the gaps in the alignment (Additional file 1: Section VI). Consequently, if the alignment between two sequences does not follow this pattern, the algorithm will not identify the alignment. In traditional dynamic programming, the alignment is obtained after computing the value of all cells in the table. However, with the banded alignment optimisation, only cells on the diameter and close to diagonal are evaluated. gl is the width of the band in banded alignment where cells farther than gl to the diameter are not evaluated. Darker cells in Fig. 1 show the case where gl=1.
Unlike traditional dynamic programming approach, MEM-Align does not have a similar table to apply banded alignment. However, we found that we can simulate the same effect by limiting the number of shift operations in the MEM extraction process. For example, if we shift the query sequence up to gl to the right and to the left, we achieve banded alignment with the band of gl. Banded-alignment reduce the complexity of MEM extraction from O(n2) to O(n.(2gl+1)) where gl is a small and fixed value. Thus, the complexity of MEM extraction is O(n) when banded alignment is applied. Also, with the said banded alignment, it is likely that fewer MEMs are extracted which speeds up the subsequent algorithmic steps.
Short MEM removal
We observed that the majority of extracted MEMs are short and are the result of randomly matching symbols. To speed up MEM-Align, MEMs shorter than sl are filtered out during MEM extraction process. This reduces the number of MEMs to be extracted and processed (subsequently speeding up the algorithm). Filtering short MEM is done by extending Algorithm 1 with a set of shift and bitwise operations (Additional file 1: Section VII).
On the other hand, there are rare cases in which short MEMs are part of the alignment; for example, a matching symbol surrounded by mismatches. Without having all MEMs in the input list, DP-MEM is not able to find the same alignment as it finds when all MEMs exist in the input list. In order to compensate for lost short MEMs in the input, we modify Φ2H of DP-MEM to consider the possibility of having short matches between MEMs (Additional file 1: Section VIII).
There might be more difficult cases where in the alignment, multiple short MEMs exist between two MEMs (see Fig. 9). The only way to correctly identify the score for the area between Mi and Mj in Φ2H is to apply a global alignment to this region. However, Φ2H is a frequent operation and should remain fast. Consequently, we decided to partially overcome the problem by limiting possible cases (a heuristic method).
If there are gaps in the area between Mi and Mj, we assume there is only one continuous gap either to the left end or to the right end of the area. Then, only two alignments are possible for the area. The number of matching symbols is counted for both cases in a sequential manner and the one that results in maximum matches is taken as the number of matches between Mi and Mj (Additional file 1: Section IX). The sequential comparison is an expensive operation and we devise a method to avoid the sequential comparison when possible (Additional file 1: Section X).
Any other case that does not fit the above assumption results in an alignment with a lower score. However, considering the low rate of gaps and mismatches, the possibility of having multiple gaps and mismatches in a small area is low.
Efficient alignment extension
In Φ2 of DP-MEM, Mj extends all alignments that end in {M1…Mj−1} (if possible). However, for each Mj there is a smaller subset Ωj⊆{M1…Mj−1} such that by extending Mj to all alignments ending in a Mi∈Ωj the alignment which ends in Mj is found (Eq. 2). In other words, there would be fewer \(S_{i}^{j}\) to be evaluated. Definition of the set Ωj and the proof of Eq. 2 are provided in Additional file 1: Section XI. The definition of Ωj is affected when short MEM removal optimisation is applied (Additional file 1: Section XII).
Hybrid alignment
To maintain the accuracy of the algorithm, we decided to utilise a hybrid method that is a combination of MEM-Align and Smith-Waterman algorithm. We define three cases in which MEM-Align may be inaccurate. If the alignment of a pair of sequences falls down into one of these cases, we use the Smith-Waterman algorithm to align sequences. These cases are:
-
When the sequences are repetitive, and the number of extracted MEMs exceeds the threshold TM. We found MEM-Align is likely to produce inaccurate alignment when aligning repetitive sequences. An appropriate TM value decreases the chance of reporting inaccurate alignment with a negligible increase in the average processing time.
-
When the computed alignment score for the alignment generated by MEM-Align is lower than a threshold TS. This case mostly occurs when there is a gap in the alignment which cannot be identified due to banded alignment.
-
When no MEM longer than sl exists to be extracted (a rare case). If sl is set to a high value and the similarity between sequences is low,
Although sending sequence pairs to an external algorithm results in additional computation, the number of sequences sent to the external algorithm remains small if appropriate values are chosen for TM and TS.
Skipping distant MEMs
When the distance between Mi and Mj is large, it is not likely to have Mi and Mj as adjacent MEMs in the alignment. Therefore, the algorithm skips the extension if the distance between Mi and Mj is longer than a threshold TD (further reducing the number of \(S_{i}^{j}\) to be evaluated). This optimisation slightly improves the performance with a negligible side effect on accuracy.
Results
In order to evaluate MEM-Align, four synthetic datasets and one realistic dataset, shown in Table 3, were considered. Each of these contains one million sequence pairs. Synthetic datasets were prepared by random selection of short sequences from the reference human genome followed by simulated variations. The realistic dataset was taken from mapped reads of sample HG00096 downloaded from the 1000 genomes project [35]. In the realistic dataset, each mapped read is paired with a sequence of the same size taken from its mapping location in the reference-genome. All datasets are available along with the MEM-Align package. These multiple datasets allowed estimating the impact of sequence length and sequence divergence levels on the speed and accuracy of the various algorithms. All the tests were run on a Linux (version 3.13.0-58-generic) machine with Intel i5-3470 processors, in single thread mode.
Two different configurations of MEM-Align (MA1 and MA2) as described in Table 4 were compared with four other alignment algorithms: A SIMD implementation of Smith-Waterman (SSW) [22]; an implementation of Ukkonen algorithm (UKK) taken from SNAP [12]; a Gene Myers algorithm (GM); and, a combination of Gene Myers with Hirschberg algorithm (GMH) implemented in the SeqAN package [23]. In order to implement hybrid alignment, MEM-Align uses the SSW algorithm internally. The appropriate values of TM and TS depend on sequence length and varies for each dataset (Table 4). The lower TS in MA2 (compared to MA1) leads to a lower number of sequence pairs being sent to SSW and results in a faster but less accurate alignment. On the other hand, MA1 delivers higher accuracy at the cost of higher execution time.
Accuracy was measured using two different metrics: the number of suboptimal alignments; and, the average alignment score difference between the reported alignment score and the optimal alignment score (reported by SSW). For the purposes of measuring accuracy, if the alignment score reported by an algorithm is less than the alignment score reported by SSW, the alignment is considered suboptimal.
Details regarding how SSW, UKK, GM and GMH were used in our experiments are available in Additional file 1: Section XIII. Execution time and accuracy are measured for existing alignment algorithms as well as for several configurations of MEM-Align, showing the effect of varying parameters on differing datasets. In order to get a deeper insight into the effect of the optimisations on MEM-Align the following metrics were also measured.
-
The number of sequences sent to SSW because of TM and TS.
-
The average number of extracted MEM for those sequences that are not sent to SSW.
-
Number of alignment extensions (number of evaluated \(S_{i}^{j}\) in Fig. 7) avoided by Ω and TD optimisation.
-
Number of times sequential string comparison of the area between Mi and Mj is avoided.
-
Proportion of execution time spent on each of the sub-processes including file access (IO), string to bit-vector conversion, MEM extraction, sorting, alignment, backtracking and the time spent by SSW to process sequence pairs sent to SSW.
The complete experimental report is provided in Additional file 1: Section XV. Here, we only highlight a fraction of the results that demonstrate the effects of various parameters. Note that for a streamlined application where the error rate, genome and sequence length are fixed, MEM-Align parameters only need to be tuned once. Thus, for the purpose of this evaluation, the MEM-Align parameters were tuned using datasets that were different from the ones used in the evaluation.
Our string to bit-vector conversion function (Additional file 1: Section III) is about 25 times faster than the conventional shift-and-insert loop method. Using the counting sort strategy to sort MEMs (Additional file 1: Section I) is 8.2 and 3.4 times faster than using the quick-sort algorithm when the average number of extracted MEMs for a pair of sequences is about 1330 and 42 respectively.
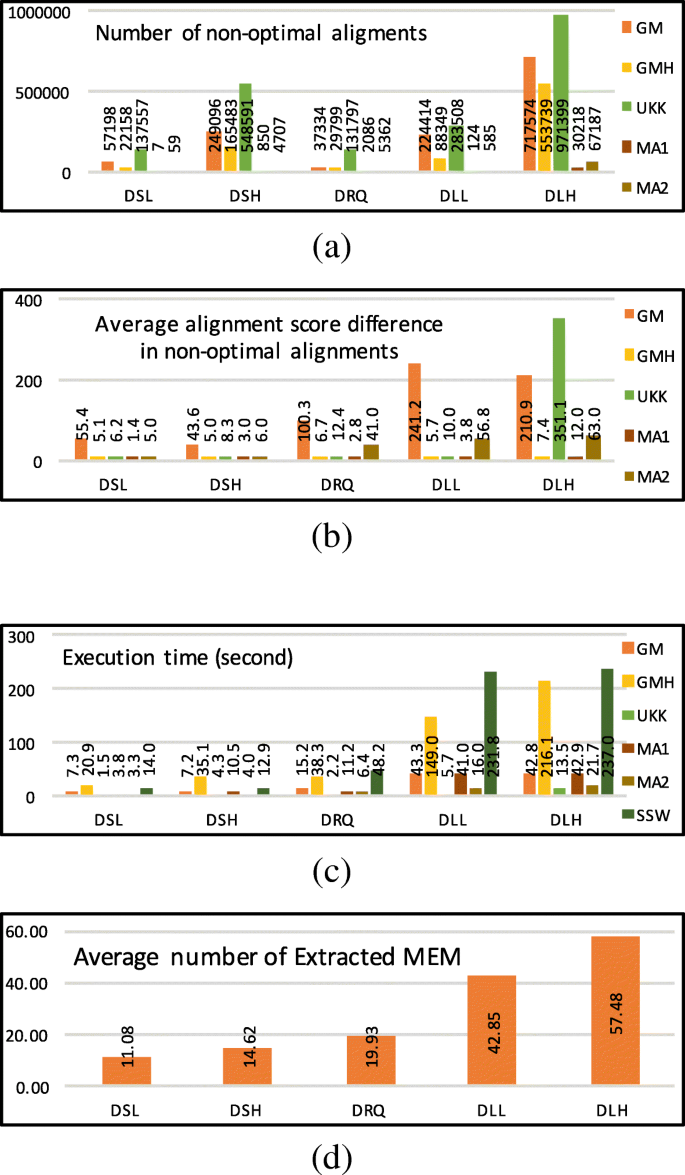
Figure 10a, b and c show the number of suboptimal alignments, average alignment score difference, and execution time respectively. While the execution time of SSW is quadratic in the length of the sequence, the UKK execution time seems to be linear in sequence length. The MEM-Align execution time is a more complex function and depends on other factors such as error rate and given parameters. Although UKK is the fastest algorithm, MEM-Align results in a considerably lower number of suboptimal alignments and is only slightly slower than UKK. Figure 10a suggests that the alignment produced by edit-distance based methods such as GM and UKK differs from the alignment produced by SSW (gold standard) for a large number of input sequences. However, MEM-Align can produce alignment identical to SSW in most cases. Thus MEM-Align is a better alternative for SSW in read-mapping applications.
Comparing two configurations of MEM-Align (MA1 and MA2) with Gene Myers (GM) and Gene Myers and Hirschberg (GMH) as well as Ukkonen (UKK) and Smith-Waterman (SSW) algorithms. a Number of suboptimal alignments. b Average alignment score difference in suboptimal alignments. c Execution time in seconds. d Average number of extracted MEMs for a pair of sequences (MA2)
Figure 10d represents the average number of extracted MEMs for a pair of sequences (Θ) for MA2 configuration of MEM-Align. Θ is in direct relation with the time spent on execution time. Θ is a helpful guideline for identifying the optimal value of TM. Although the exact function has not been identified yet, it appears that 4Θ<TM<5Θ is a suitable estimation.
The proportion of execution time for each algorithmic step of MEM-Align is shown in Fig. 11a. Since the execution times for some configuration of MEM-Align are negligible compared to other configurations, in Fig. 11a all execution times are scaled to a 100% bar to allow for better visualisation. MA1 spends more time in SSW since more sequences are sent to SSW because of higher value for TS.
The effect of proposed optimisations on MEM-Align. a Proportion of execution time for different algorithmic steps. b Number of sequence pairs sent to SSW by TM and TS. c Number of alignment extensions in million for MA2 (normalised bar). d Number of string comparisons per million for MA2 (normalised bar). e Execution times varying gl and sl. f Number of suboptimal alignments varying gl and sl
The number of sequence pairs sent to SSW because of TM and TS are shown in Fig. 11b. Data in Fig. 11b are scaled to 100% bar in order to reflect the percentage of sequence pairs sent to SSW yet the labels show the actual value.
Figure 11c reports the expected number of times the alignment extension is executed along with the number of times alignment extension is avoided (optimised) because of using the set Ω and the parameter TD. Both of these optimisations have been noticeably effective. Figure 11d demonstrates the number of times sequential string comparison of the area between Mi and Mj is avoided. All bars are scaled to 100%, yet labels show the actual number in millions.
The effect of varying sl and gl (on DLL dataset) are shown in Fig. 11e (execution time) and Fig. 11f (the number of suboptimal alignments). While sl<4 exponentially increases execution time, sl>4 result in significant increase in number of suboptimal alignments. sl=4 seem to be the most appropriate value. gl=6 delivers the best trade off between speed and accuracy for this data set. In these figures TM=1,000,000, TD=1,000 and TS=0 are set to disable corresponding optimisation and only show the effect of sl and gl. To see the effect of TM, TS and TD refer to Additional file 1: Section XV.
Discussion
Due to the similarity in many operations, there is potential to internally implement the SHD filter [36] in the MEM extraction step with minimal additional computation. In [37], SHD is accelerated using custom hardware resulting in up to 17.3 times speed up. Similar Hardware acceleration can be applied to our MEM extraction process.
Finally, we are aware that Gene Myers and Ukkonen algorithms are edit-distance based alignments and do not support affine-gap scoring. The reason we compare them to MEM-Align is that they are used in recent DNA read-mappers as an alternative to the Smith-Waterman algorithm. Our results demonstrate that MEM-Align is likely to be a better substitute for the Smith-Waterman algorithm as it allows for affine-gap scoring.
The implementation of MEM-Align contains a module that prints out human-readable colourful alignments. For each alignment, this module highlights extracted MEMs in the alignment as well as removed short MEMs which are part of the alignment and are identified by the sequential string compare operation. A sample output is provided in Additional file 1: Section XIV.
Conclusions
Pairwise alignment is one of the most frequently utilized operations in sequence analysis. Considering the growth in the amount of sequence data to be processed in the near future [38], even a small improvement in the alignment operation can save significant computational power. MEM-Align delivers considerable speed improvement, especially in the case of longer sequences where the traditional alignment methods slow down quickly.
Decision making based on sequenced data is life critical. However, errors are common in the sequencing process. Most analysis overcome errors by utilising redundant data (overlap sequences). We believe the negligible number of suboptimal alignments produced by MEM-Align can be partly compensated by the existing redundancy in the data. When speed matters, MEM-Align is the best algorithm as it is fast and its output is near identical to the Smith-Waterman algorithm with affine-gap scoring. Other alternative such as UKK and GM are also fast but only support edit-distance based scoring.
Abbreviations
- DLH:
-
Dataset of long sequences with high error rate
- DLL:
-
Dataset of long sequences with low error rate
- DRQ:
-
Dataset of real query sequences
- DSH:
-
Dataset of short sequences with high error rate
- DSL:
-
Dataset of short sequences with low error rate
- MEM:
-
Maximal exact matches
- SHD:
-
Shifted hamming distance
- SIMD:
-
Single instruction multiple data
- SSE:
-
Streaming SIMD extensions
References
Rosenberg MS. Sequence alignment: methods, models, concepts, and strategies. London: University of California Press; 2009.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009; 25(14):1754–60.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013:. p. 3. https://arxiv.org/abs/1303.3997.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10(3):25.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; 9(4):357–9.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990; 215(3):403–10.
Kent WJ. Blat—the blast-like alignment tool. Genome Res. 2002; 12(4):656–64.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004; 5(2):12.
Ma B, Tromp J, Li M. Patternhunter: faster and more sensitive homology search. Bioinformatics. 2002; 18(3):440–5.
Li M, Ma B, Kisman D, Tromp J. Patternhunter ii: Highly sensitive and fast homology search. J Bioinforma Comput Biol. 2004; 2(03):417–39.
Ferragina P, Manzini G. An experimental study of a compressed index. Inf Sci. 2001; 135(1-2):13–28.
Zaharia M, Bolosky WJ, Curtis K, Fox A, Patterson D, Shenker S, Stoica I, Karp RM, Sittler T. Faster and More Accurate Sequence Alignment with SNAP. 2011. arXiv.
Marco-Sola S, Sammeth M, Guigó R, Ribeca P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012; 9(12):1185–8.
Keich U, Li M, Ma B, Tromp J. On spaced seeds for similarity search. Discret Appl Math. 2004; 138(3):253–63.
Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970; 48(3):443–53.
Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981; 147(1):195–7.
Bille P. A survey on tree edit distance and related problems. Theor Comput Sci. 2005; 337(1-3):217–39.
Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol. 1982; 162(3):705–8.
Liu Y, Popp B, Schmidt B. CUSHAW3: sensitive and accurate base-space and color-space short-read alignment with hybrid seeding. PLoS ONE. 2014; 9(1):86869.
Farrar M. Striped Smith-Waterman speeds database searches six times over other SIMD implementations. Bioinformatics. 2007; 23(2):156–61.
Szalkowski A, Ledergerber C, Krähenbühl P, Dessimoz C. SWPS3 - fast multi-threaded vectorized Smith-Waterman for IBM Cell/B.E. and x86/SSE2. BMC Res Notes. 2008; 1:107.
Zhao M, Lee ea. SSW Library: An SIMD Smith-Waterman C/C++ Library for Use in Genomic Applications. PLoS ONE. 2013; 8(12):82138.
Döring A, Weese ea. SeqAn An efficient, generic C++ library for sequence analysis. BMC Bioinformatics. 2008; 9(1):11.
Myers G. A fast bit-vector algorithm for approximate string matching based on dynamic programming. J ACM. 1999; 46(3):395–415.
Ukkonen E. Algorithms for approximate string matching. Inf Control. 1985; 64(1):100–18.
Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning dna sequences. J Comput Biol. 2000; 7(1-2):203–14.
Liu Y, Wirawan A, Schmidt B. CUDASW++ 3.0: accelerating Smith-Waterman protein database search by coupling CPU and GPU SIMD instructions. BMC Bioinformatics. 2013; 14:117.
Harris B, Jacob AC, Lancaster JM, Buhler J, Chamberlain RD. A Banded Smith-Waterman FPGA Accelerator for Mercury BLASTP. In: 2007 International Conference on Field Programmable Logic and Applications. IEEE: 2007. p. 765–9. https://doi.org/10.1109/FPL.2007.4380764.
Allred J, Coyne J, Lynch W, Natoli V, Grecco J, Morrissette J. Smith-Waterman implementation on a FSB-FPGA module using the Intel Accelerator Abstraction Layer. In: 2009 IEEE International Symposium on Parallel & Distributed Processing. IEEE: 2009. p. 1–4. https://doi.org/10.1109/IPDPS.2009.5161214.
DePristo MA, Banks ea. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43(5):491–8.
Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, Wilkie AOM, McVean G, Lunter G, WGS500 Consortium and others. Integrating mapping-, assembly-and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014; 46(8):912.
Khan Z, Bloom JS, Kruglyak L, Singh M. A practical algorithm for finding maximal exact matches in large sequence datasets using sparse suffix arrays. Bioinformatics. 2009; 25(13):1609–16.
Aluru S. Handbook of computational molecular biology.Chapman & All/Crc Computer and Information Science Series; 2005.
Chao K-M, Pearson WR, Miller W. Aligning two sequences within a specified diagonal band. Bioinformatics. 1992; 8(5):481–7.
1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015; 526(7571):68–74. https://doi.org/10.1038/nature15393.
Xin H, Greth J, Emmons J, Pekhimenko G, Kingsford C, Alkan C, Mutlu O. Shifted Hamming distance: a fast and accurate SIMD-friendly filter to accelerate alignment verification in read mapping. Bioinformatics (Oxford, England). 2015; 31(10):1553–60.
Alser M, Hassan H, Xin H, Ergin O, Mutlu O, Alkan C. GateKeeper : Enabling Fast Pre-Alignment in DNA Short Read Mapping with a New Streaming Accelerator Architecture. 2016. http://arxiv.org/abs/1604.01789.
Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. Big data: astronomical or genomical?. PLoS Biol. 2015; 13(7):1002195.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
MEM-Align source code and datasets along with scripts and tools to replicate our experiments as well as online one-click sample run are publicly available at https://sites.google.com/site/memalignv1.
Author information
Authors and Affiliations
Contributions
AB conceived and implemented the method, prepared and ran the experiments, and wrote the manuscript. BG contribute to the biological aspects and to the writing of the manuscript. AI contributed to the algorithmic development and to the writing of the manuscript. SP supervised the project and contributed to the writing of the manuscript. The paper was written by all authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1
Supplementary Data. Supporting material as well as the complete experimental result are provided in supplementary data. (PDF 3044 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Bayat, A., Gaëta, B., Ignjatovic, A. et al. Pairwise alignment of nucleotide sequences using maximal exact matches. BMC Bioinformatics 20, 261 (2019). https://doi.org/10.1186/s12859-019-2827-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-019-2827-0